#
Onco Query Language (OQL)
#
Introduction to OQL
The Onco Query Language (OQL) is used to define which specific types of alterations are included in a query on the cBioPortal for Cancer Genomics. By default, querying for a gene includes mutations, fusions, amplifications and deep deletions. OQL can be used to specify specific mutations (e.g. BRAF V600E) or types of mutations (e.g. BRCA1 truncating mutations), lower level copy number alterations (e.g. CDKN2A shallow deletions), changes in mRNA or protein expression, and more.
OQL-specified alterations will be reflected on most tabs, including OncoPrint, but are not currently reflected on the Plots, Co-Expression or Expression tabs.
Note that OQL assumes any word that it doesn't recognize is a mutation code.
Additional explanation and examples using OQL are available in the User Guide.
#
OQL Keywords
Users can define specific subsets of genetic alterations for five data types:
* These are the default OQL keywords used for each data type when a gene is queried without any explicit OQL.
#
OQL modifiers
Mutations and copy number alterations can be further refined using modifiers:
#
Basic Usage
When querying a gene without providing any OQL specifications, cBioPortal will default to these OQL terms for a query with Mutation and Copy Number selected in the Genomic Profiles section:
MUT FUSION AMP HOMDEL
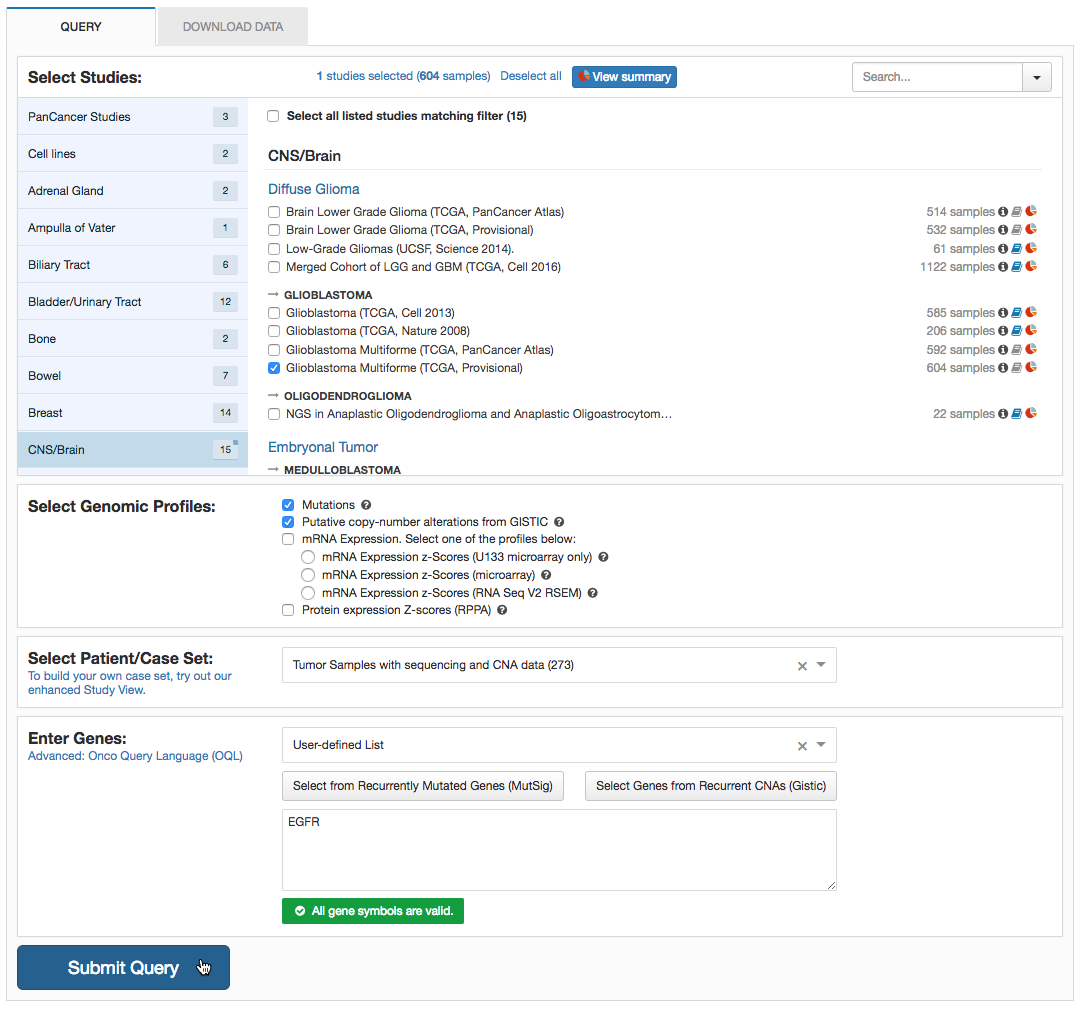
You can see the OQL terms applied by hovering over the gene name in OncoPrint:
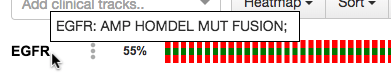
If you select RNA and/or Protein in the "Genomic Profiles" section of the query, the default settings are:
RNA: EXP >= 2 EXP <= -2
Protein: PROT >= 2 PROT <= -2
You must select the relevant Genomic Profile in order for OQL to query that data type. For example, you can't add EXP > 2 to the query without also selecting an RNA profile.
Proper formatting for OQL is straightforward: gene name, followed by a colon, followed by any OQL keywords and ending in a semicolon, an end-of-line, or both.
GENE1: OQL KEYWORDS;
GENE2: OQL KEYWORDSIn general, any combination of OQL keywords and/or expressions can annotate any gene, and the order of the keywords is immaterial.
Below we will go into greater detail about each data type.
#
Mutations
To view cases with specific mutations, provide the specific amino acid change of interest:
BRAF: MUT = V600EYou can also view all mutations at a particular position:
BRAF: MUT = V600Or all mutations of a specific type:
TP53: MUT = <mutation type><mutation type> can be one or more of:
MISSENSENONSENSENONSTARTNONSTOPFRAMESHIFTINFRAMESPLICETRUNC
For example, to view TP53 truncating mutations and in-frame insertions/deletions:
TP53: MUT = TRUNC INFRAMEOQL for mutations can also be written without MUT =. The following examples are identical:
BRAF: MUT = V600E
BRAF: V600E
TP53: MUT = TRUNC INFRAME
TP53: TRUNC INFRAMEOQL can also be used to exclude a specific protein change, position or type of mutation. For example, below are examples to query all EGFR mutations except T790M, all BRAF mutations except those at V600 and all TP53 mutations except missense:
EGFR: MUT != T790M
BRAF: MUT != V600
TP53: MUT != MISSENSENote that this will only work to exclude a single event. Because OQL uses 'OR' logic, excluding multiple mutations or excluding a mutation while including another mutation (e.g. BRAF: MUT=V600 MUT!=V600E) will result in querying all mutations.
#
Copy Number Alterations
To view cases with specific copy number alterations, provide the appropriate keywords for the copy number alterations of interest. For example, to see amplifications:
CCNE1: AMPOr amplified and gained cases:
CCNE1: CNA >= GAINWhich can also be written as:
CCNE1: GAIN AMP
#
Expression
High or low mRNA expression of a gene is determined by the number of standard deviations (SD) from the mean. For example, to see cases where mRNA for CCNE1 is greater than 3 SD above the mean:
CCNE1: EXP > 3
#
Protein
High or low protein expression is similarly determined by the number of SD from the mean. For example, to see cases where protein expression is 2 SD above the mean:
EGFR: PROT > 2Protein expression can also be queried at the phospho-protein level:
EGFR_PY992: PROT > 2
#
Modifiers
Modifiers can be used on their own or in combination with other OQL terms for mutations, fusions and copy number alterations to further refine the query. Modifiers can be combined with other OQL terms using an underscore. The order in which terms are combined is immaterial.
#
Driver
The DRIVER modifier applies to mutations, fusions and copy number alterations. The definition of what qualifies as a driver alteration comes from the "Mutation Color" menu in OncoPrint. By default, drivers are defined as mutations, fusions and copy number alterations in OncoKB or CancerHotspots.
On its own, the DRIVER modifier includes driver mutations, fusions and copy number alterations:
EGFR: DRIVEROr it can be used in combination with another OQL term. For example, to see only driver fusion events:
EGFR: FUSION_DRIVEROr driver missense mutations:
EGFR: MUT = MISSENSE_DRIVERWhen combining DRIVER with another OQL term, the order doesn't matter: MUT_DRIVER and DRIVER_MUT are equivalent. DRIVER can be combined with:
MUTMUT = <mutation type>orMUT = <protein change>FUSIONCNAAMPorGAINorHETLOSSorHOMDELGERMLINEorSOMATIC(see below)
#
Germline/Somatic
The GERMLINE and SOMATIC modifiers only apply to mutations. A mutation can be explicitly defined as germline during the data curation process. Note that very few studies on the public cBioPortal contain germline data.
GERMLINE or SOMATIC can be combined with:
MUTMUT = <mutation type>orMUT = <protein change>DRIVER
To see all germline BRCA1 mutations:
BRCA1: GERMLINEOr to see specifically truncating germline mutations:
BRCA1: TRUNC_GERMLINE
BRCA1: GERMLINE_TRUNCThe order is immaterial; both options produce identical results.
Or to see somatic missense mutations:
BRCA1: MUT = MISSENSE_SOMATICGERMLINE or SOMATIC can also be combined with DRIVER and, optionally, a more specific mutation term (e.g. NONSENSE):
BRCA1: NONSENSE_GERMLINE_DRIVER
#
The DATATYPES Command
To save copying and pasting, the DATATYPES command sets the genetic annotation for all subsequent genes. Thus,
DATATYPES: AMP GAIN HOMDEL EXP > 1.5 EXP < -1.5; CDKN2A MDM2 TP53is equivalent to:
CDKN2A: AMP GAIN HOMDEL EXP > 1.5 EXP < -1.5
MDM2: AMP GAIN HOMDEL EXP > 1.5 EXP < -1.5
TP53: AMP GAIN HOMDEL EXP > 1.5 EXP < -1.5
#
Merged Gene Tracks
OQL can be used to create a merged gene track in OncoPrint, in which alterations in multiple genes appear as a single track. This is done by enclosing a list of genes in square brackets. By default, the track will be labeled by the gene names, separated by '/'. To instead specify a label, type the desired label within double quotes at the beginning of the square brackets. For example:
["CDK INHIBITORS" CDKN2A CDKN2B]
[MDM2 MDM4]The resulting merged gene track will be visible in OncoPrint and can be expanded to view the individual gene tracks. For example:
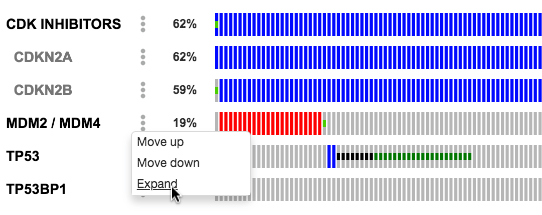
https://www.cbioportal.org/results/oncoprint?session_id=5c1966e2e4b05228701f958e
It is possible to include OQL for specific alterations in merged gene tracks, as well as querying a combination of single and merged gene tracks.
Note that merged gene tracks only appear in OncoPrint. All other pages show the individual genes.
#
Example: RB Pathway Alterations
Provided below is one example of the power of using OQL. Additional examples are available in the User Guide.
#
Using the Defaults
Select Ovarian Serous Cystadenocarcinoma (TCGA, Nature 2011) with the following data types:
- Mutations
- Putative copy-number alterations (GISTIC)
- mRNA expression (mRNA expression Z-scores (all genes))
Input the following three genes in the RB pathway:
- CCNE1
- RB1
- CDKN2A
Submit this query and note how many samples have alterations in multiple of these genes:
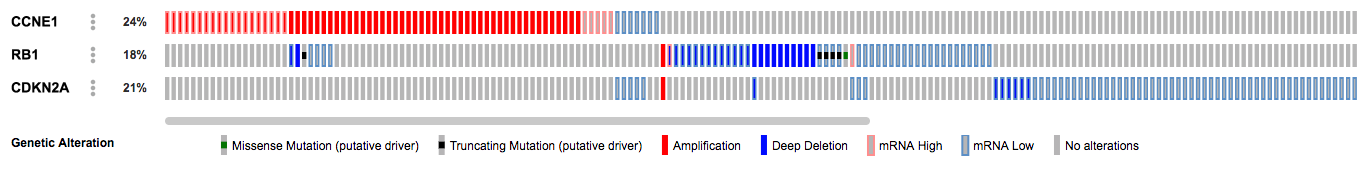
https://www.cbioportal.org/results/oncoprint?session_id=5c1966cee4b05228701f958d
#
Greater Insight with OQL
Given what is known about the RB pathway, the events that are most likely selected for in the tumors are CCNE1 amplification, RB1 deletions or mutations, and loss of expression of CDKN2A. To investigate this hypothesis, we can use OQL to display only these events. Modify the query to reflect this:
CCNE1: AMP MUT
RB1: HOMDEL MUT
CDKN2A: HOMDEL EXP < -1Examine the updated OncoPrint:
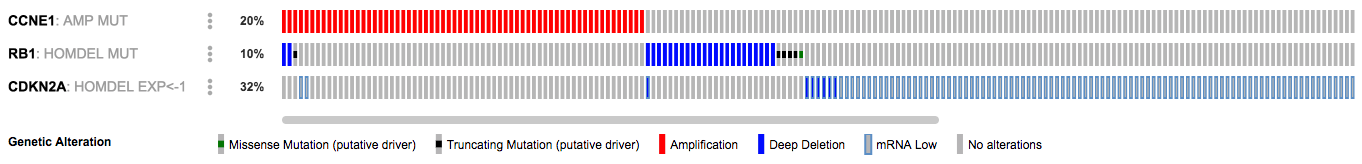
https://www.cbioportal.org/results/oncoprint?session_id=5c1966aee4b05228701f958c
This shows that alterations in these genes are almost entirely mutually-exclusive -- no cases are altered in all three genes and only six are altered in two genes. This supports the theory that the tumor has selected for these events.
#
Questions? Feedback?
Please share any questions or feedback on OQL with us: https://groups.google.com/group/cbioportal
Also note that additional explanation and examples using OQL are available in the User Guide.
