#
News
#
September 27, 2024
- Added data consisting of 2,409 samples from 10 studies:
- Pediatric European MAPPYACTS Trial (Gustave Roussy, Cancer Discov 2022) 786 samples
- Hepatocellular Carcinoma (CLCA, Nature 2024) 494 samples
- Gastrointestinal Stromal Tumors (MSK, Clin Cancer Res 2023) 469 samples
- Diffuse Large B-Cell Lymphoma (MSK, Haematologica 2024) 396 samples
- Pituitary Adenoma (MSK, Acta Neuropathologica 2024) 104 samples
- Triple-Negative Breast Cancer (FUSCC, Cell Research 2020) 69 samples
- Endometrial and Ovarian Cancer (MSK, Nature Medicine 2024) 33 samples
- Cutaneous Squamous Cell Carcinoma (UOW, Front Oncol 2022) 25 samples
- RAD51B Associated Mixed Cancers (MSK, NPJ Breast Cancer 2021) 19 samples
- Pleural Mesothelioma (MSK, Clin Cancer Res 2024) 14 samples
#
August 28, 2024
- New feature (Windows users only): The Group Comparison results can now be visualized in 3D with AVM, an interactive 3D visualization software tool with data shaping functions and integrated pathways. Read more on AVM for cBioPortal.
- Note for private instances: To enable this feature in private cBioPortal instances, please see this instruction.
- Note for private instances: To enable this feature in private cBioPortal instances, please see this instruction.
#
July 29, 2024
- Added data consisting of 12,863 samples from 33 TCGA and 10 CPTAC studies from the Genomic Data Commons (GDC) as part of the Cancer Research Data Commons (NCI-CRDC) initiative. More information can be found on our FAQ.
- Acute Myeloid Leukemia (TCGA, GDC) 200 samples
- Adrenocortical Carcinoma (TCGA, GDC) 92 samples
- Bladder Urothelial Carcinoma (TCGA, GDC) 413 samples
- Cervical Squamous Cell Carcinoma (TCGA, GDC) 309 samples
- Cholangiocarcinoma (TCGA, GDC) 51 samples
- Chromophobe Renal Cell Carcinoma (TCGA, GDC) 66 samples
- Colon Adenocarcinoma (TCGA, GDC) 463 samples
- Cutaneous Melanoma (TCGA, GDC) 473 samples
- Diffuse Glioma (TCGA, GDC) 530 samples
- Diffuse Large B-Cell Lymphoma, NOS (TCGA, GDC) 48 samples
- Endometrial Carcinoma (TCGA, GDC) 548 samples
- Esophageal Adenocarcinoma (TCGA, GDC) 186 samples
- Glioblastoma Multiforme (TCGA, GDC) 610 samples
- Head and Neck Squamous Cell Carcinoma (TCGA, GDC) 530 samples
- Hepatocellular Carcinoma (TCGA, GDC) 379 samples
- High-Grade Serous Ovarian Cancer (TCGA, GDC) 602 samples
- Invasive Breast Carcinoma (TCGA, GDC) 1103 samples
- Lung Adenocarcinoma (TCGA, GDC) 571 samples
- Lung Squamous Cell Carcinoma (TCGA, GDC) 503 samples
- Miscellaneous Neuroepithelial Tumor (TCGA, GDC) 184 samples
- Non-Seminomatous Germ Cell Tumor (TCGA, GDC) 156 samples
- Pancreatic Adenocarcinoma (TCGA, GDC) 186 samples
- Papillary Renal Cell Carcinoma (TCGA, GDC) 292 samples
- Papillary Thyroid Cancer (TCGA, GDC) 515 samples
- Pleural Mesothelioma (TCGA, GDC) 87 samples
- Prostate Adenocarcinoma (TCGA, GDC) 501 samples
- Rectal Adenocarcinoma (TCGA, GDC) 171 samples
- Renal Clear Cell Carcinoma (TCGA, GDC) 537 samples
- Soft Tissue Cancer (TCGA, GDC) 264 samples
- Stomach Adenocarcinoma (TCGA, GDC) 443 samples
- Thymoma (TCGA, GDC) 124 samples
- Uterine Carcinosarcoma/Uterine Malignant Mixed Mullerian Tumor (TCGA, GDC) 57 samples
- Uveal Melanoma (TCGA, GDC) 80 samples
- Breast Cancer (CPTAC, GDC) 154 samples
- CNS/Brain Cancer (CPTAC, GDC) 74 samples
- Colon Adenocarcinoma (CPTAC, GDC) 109 samples
- Head and Neck Carcinoma, Other (CPTAC, GDC) 150 samples
- Lung Adenocarcinoma (CPTAC, GDC) 180 samples
- Lung Squamous Cell Carcinoma (CPTAC, GDC) 90 samples
- Ovarian Cancer (CPTAC, GDC) 113 samples
- Pancreatic Cancer (CPTAC, GDC) 139 samples
- Renal Cell Carcinoma (CPTAC, GDC) 288 samples
- Uterine Endometrioid Carcinoma (CPTAC, GDC) 292 samples
#
July 10, 2024
- New Feature: Explore mutational signatures on the patient page. The Pan-cancer analysis of whole genomes (ICGC/TCGA, Nature 2020) study has mutational signatures data available for over 2,500 cases. The new Mutational Signatures tab on the patient page graphically displays the types of mutations found in each case along with COSMIC reference signatures. Example: POLE signature in case DO8898 in Pan-cancer analysis of whole genomes (ICGC/TCGA, Nature 2020)
image
#
May 29, 2024
Added data consisting of 9,632 samples from 10 studies:
- Soft Tissue and Bone Sarcoma (MSK, Nat Commun 2022) 7494 samples
- Chronic Lymphocytic Leukemia (Broad, Nature Genetics 2022) 1154 samples
- Mature B-Cell Neoplasms (Simon Fraser University, Blood 2023) 297 samples
- Anaplastic Thyroid Cancers (GATCI, Cell Reports 2024) 190 samples
- Colorectal Cancer (CAS Shanghai, Cancer Cell 2020) 146 samples
- Prostate Cancer MDA PCa PDX (MD Anderson, Clin Cancer Res 2024) 88 samples
- Ovarian Cancer - MSK SPECTRUM (MSK, Nature 2022) 82 samples
- IDH-mutated Diffuse Glioma (MSK, Clin Cancer Res 2024) 73 samples
- Pre-cancer Colorectal Polyps (HTAN Vanderbilt, Cell 2021) 61 samples
- Prostate Cancer (MSK, Science 2022) 47 samples
Data Improvement
- Pan-can studies Ancestry addition: Genetic Ancestry data is added to all 32 TCGA Pan-Can studies,in generic assay format. Data source: GDC. Example: Ancestry-associated somatic genetic alterations
- Pan-can studies Ancestry addition: Genetic Ancestry data is added to all 32 TCGA Pan-Can studies,in generic assay format. Data source: GDC. Example: Ancestry-associated somatic genetic alterations
#
May 7, 2024
New Feature: Gene-specific charts in Study View now support mutation data. Two chart types are available: sample mutational status and mutation types.
New Feature: Categorical data loaded into the generic assay format is now available in group comparison. Example: Arm-level CNA compared between astrocytoma vs oligodendroglioma in Brain Lower Grade Glioma (TCGA, PanCancer Atlas)
Enhancement: OncoPrint now allows you to remove a gene track.
#
Apr 2, 2024
- Introducing the cBioPortal Newsletter! Stay updated with the latest developments, insights, and community highlights of cBioPortal. Subscribe via LinkedIn or google groups. We'll be sharing valuable updates every few months.
#
March 27, 2024
New Feature: The Plots tab is now available in the Study View. After exploring a cohort and applying any filters of interest in the Study View Summary page, you can now click over to the new Plots tab to explore the cohort by plotting any two attributes against each other. Example: Mutation count vs subtype in Uterine Corpus Endometrial Carcinoma (TCGA, Nature 2013)
Enhancement: The custom selection feature in Study View, which filters to a user-defined list of samples/patients, no longer requires the inclusion of the study ID with each sample ID. If viewing multiple studies in study view and filtering to a sample ID which exists in more than one study, all samples will be displayed by default or the study ID can be included for additional specificity.
#
Mar 5, 2024
- Local Installations Feature: Make representation of custom driver annotation configurable:
#
Feb 7, 2024
New feature: The Datasets Page now lists what studies have samples profiled for Structural Variants:
New Major Release: v6.0.0 includes a major repackaging of the backend software to improve the development experience. The backend has been upgraded to use Java Spring Boot v3.1.4. The CORE and MAF modules have been moved to new repositories. All other modules have been compacted into a single source repository. We have also updated the JVM to 21 and many libraries have been updated to address security and performance issues. See more information in the v6.0.0 release notes.
Local Installations Feature: When mutational signature data is loaded, show COSMIC reference signatures on the Patient View:
#
Dec 29, 2023
Added data consisting of 5,120 samples from 13 studies:
- Endometrial Cancer (MSK, Cancer Discovery 2023) 1882 samples
- Esophagogastric Cancer (MSK, J Natl Cancer Inst 2023) 902 samples
- Diffuse Glioma (GLASS Consortium, 2022) 693 samples
- Bladder Cancer (MSK, Clin Cancer Res 2023) 526 samples
- Non-Small Cell Lung Cancer Brain Metastasis (MSK, Nat Commun 2023) 322 samples
- MSK Make-an-IMPACT Rare Cancers (MSK, Clin Cancer Res 2023) 184 samples
- Cervical Cancer (MSK, Clin Cancer Res 2023) 177 samples
- Mature T and NK Neoplasms (MSK, Blood Adv 2023) 132 samples
- Hepatocellular Carcinoma (MSK, JCO Precis Oncol 2023) 90 samples
- Sarcoma (MSK, J Pathol 2023) 82 samples
- Esophagogastric Cancer (MSK, Clin Cancer Res 2023) 64 samples
- Rhabdomyosarcomas (MSK, NPJ Precis Oncol 2023) 42 samples
- Pediatric Rhabdomyosarcomas (MSK, JCO Precis Oncol 2023) 24 samples
Gene Tables Update: Updated tables of genes (main and alias), based on Oct 1, 2023 HGNC release. See seedDB release notes here for details.
#
Dec 5, 2023
New feature: Filter by data types on the homepage:
New feature: Change colors of tracks in Oncoprint:
New feature: Show alteration frequencies per group in OncoPrint. Example: RTK-RAS alterations in Smokers vs Never Smokers Lung Cancer Cases in MSK-IMPACT Clinical Sequencing Cohort
#
Oct 17, 2023
New feature: Survival charts with landmark events and hazard ratios. Example: TP53 in Lung Cancer Cases in MSK-IMPACT Clinical Sequencing Cohort
#
Oct 3, 2023
New feature: New Structural Variants Tab on Results View. Example: TMPRSS2 Structural Variants in MSK-IMPACT Clinical Sequencing Cohort
#
Sep 5, 2023
New feature: Add gene-specific CNA charts to show all levels of copy number alterations (including gain and hetloss) on Study View:
#
Aug 30, 2023
Enhancement: Exclude a patient from your selection directly from the Patient View:
#
Aug 21, 2023
Added data consisting of 4,488 samples from 7 studies:
- Lung Adenocarcinoma Met Organotropism (MSK, Cancer Cell 2023) 2653 samples
- Acute Myeloid Leukemia (OHSU, Cancer Cell 2022) 942 samples
- Colon Cancer (Sidra-LUMC AC-ICAM, Nat Med 2023) 348 samples
- Pediatric Neuroblastoma (MSK, Nat Genet 2023) 223 samples
- Colorectal Adenocarcinoma (MSK, Nat Commun 2022) 180 samples
- Bladder Cancer (Columbia University/MSK, Cell 2018) 130 samples
- Myoepithelial Carcinomas of Soft Tissue (WCM, CSH Molecular Case Studies 2022) 12 samples
Gene Tables Update: Updated tables of genes (main and alias), based on Apr 1, 2023 HGNC release. See seedDB release notes here for details.
#
Aug 1, 2023
Enhancement: One-sided Fisher's exact tests were changed to be two-sided. The affected pages are:
- Results View Page - Mutual Exclusivity Tab
- Results View Page - Comparison Tab - Genomic Alterations Tab
- Comparison Page - Genomic Alterations Tab
- Comparison Page - Mutations Tab
Please note that the Mutations tab on the Comparison page is a recent feature and was introduced with the two-sided Fisher's exact test already implemented.
Several users pointed out that using a one-sided test was incorrect for these comparisons. Please see discussions here for more information.
#
Jul 18, 2023
New Feature: Add mutations table and two-sided exact p-value to comparison. Example: AR mutations in Primary vs Metastatic Prostate Cancer samples in MSK-IMPACT Clinical Sequencing Cohort
#
May 2, 2023
New Feature: The mutations tab now shows variant annotations from the repository of Variant with Unexpected Effects (reVUE).

#
Apr 25, 2023
- Local Installations Feature: When custom driver annotations for structural variants are loaded, one can now filter by them in both the Oncoprint and the Study View.
#
Apr 11, 2023
New Feature: Disable autocommit and manually commit filters in study view. Manually commit filters can improve cBioPortal performance when query large dataset.


#
Apr 5, 2023
Added data consisting of 2,472 samples from 5 studies:
- Bladder Cancer (MSK, Cell Reports 2022) 1659 samples
- Gastrointestinal Stromal Tumor (MSK, NPJ Precis Oncol 2023) 499 samples
- Appendiceal Cancer (MSK, J Clin Oncol 2022) 273 samples
- Colorectal Cancer (MSK, Cancer Discovery 2022) 22 samples
- Nerve Sheath Tumors (Johns Hopkins, Sci Data 2020) 19 samples [First GRCh38 Study]
Data Improvement
- Added TERT promoter mutation status to Melanomas (TCGA, Cell 2015), Papillary Thyroid Carcinoma (TCGA, Cell 2014) TCGA studies.
#
Apr 4, 2023
New Feature: Allow numeric data type for custom data charts.


This also allows to have numerical custom data after we query based on genes (custom data 2 in the image):


#
Jan 10, 2023
New Feature: New Pathways tab on the Group Comparison view. Example: Primary vs Metastasis samples in MSK-IMPACT Clinical Sequencing Cohort

#
Dec 13, 2022
New Feature: New Mutations tab on the Group Comparison view. Example: Primary vs Metastasis samples in MSK-IMPACT Clinical Sequencing Cohort

#
Oct 12, 2022
- Added data consisting of 1,459 samples from 10 studies:
- Hepatocellular Carcinoma (MERiC/Basel, Nat Commun. 2022) 122 samples
- Prostate Cancer Brain Metastases (Bern, Nat Commun. 2022) 168 samples
- Pan-Cancer MSK-IMPACT MET Validation Cohort (MSK 2022) 69 samples
- Endometrial Carcinoma cfDNA (MSK, Clin Cancer Res 2022) 44 samples
- Endometrial Carcinoma MSI (MSK, Clin Cancer Res 2022) 181 samples
- Gallbladder Cancer (MSK, Clin Cancer Res, 2022) 244 samples
- Meningioma (University of Toronto, Nature 2021) 121 samples
- Mixed Tumors: Selpercatinib RET Trial (MSK, Nat Commun. 2022) 188 samples
- Low-Grade Serous Ovarian Cancer (MSK, Clin Cancer Res 2022) 119 samples
- Urothelial Carcinoma (BCAN/HCRN 2022) 203 samples
#
Sep 6, 2022
Enhancement: Oncoprint can now save clinical tracks after login:

#
Aug 11, 2022
- New Major Release: v5.0.0 release drops support for fusions in the mutation data format. Going forward fusions can only be imported in the Structural Variant (SV) format. This is mainly a refactoring effort to simplify the codebase and pave the way for the development of novel structural variant visualizations in the future. For cBioPortal instance maintainer, please reference our Migration Guide for instruction.
#
Jul 26, 2022
- Added data consisting of 6,631 samples from 7 studies:
- Metastatic Biliary Tract Cancers (SUMMIT - Neratinib Basket Trial, 2022) 36 samples
- Rectal Cancer (MSK, Nature Medicine 2022) 801 samples
- Lung Adenocarcinoma (MSK Mind,Nature Cancer 2022) 247 samples
- Myelodysplastic Syndromes (MDS IWG, IPSSM, NEJM Evidence 2022) 3,323 samples
- Esophagogastric Cancer (MSK, Clin Cancer Res 2022) 237 samples
- Pan-cancer Analysis of Advanced and Metastatic Tumors (BCGSC, Nature Cancer 2020) 570 samples
- Prostate Adenocarcinoma (MSK, Clin Cancer Res. 2022) 1,417 samples
#
Jun 7, 2022
New Feature: Add heatmap to plot options on Comparison Page. Example: Primary vs Metastatic Prostate Cancer in MSK-IMPACT (2017)
#
May 31, 2022
New Feature: Added Quartiles, Median split and Generate bins options for bar charts on the study view page, where Generate bins allows user to define bin size and min value
#
May 12, 2022
New Feature: Show cohort alteration frequencies in pathways from NDEx on the Results View. Example: Glioblastoma signaling pathways in MSK-IMPACT (2017) cohort

#
May 24, 2022
- New Feature: Add Help buttons on various pages and tabs, including the homepage
#
May 10, 2022
New Feature: Use IGV for the genomic overview on the Patient View. Example: Endometrial cancer patient in TCGA
#
May 5, 2022
New Feature: View mutations and copy number changes in the Integrative Genomics Viewer (IGV) on the Patient View. Example: Endometrial cancer patient in TCGA

New Feature: Add charts that plot categorical vs continuous data on the Study View. Example: MSK-IMPACT (2017) cohort

New Feature: Several single cell data integrations are now available for the CPTAC glioblastoma study, allowing one to:
- Compare genomic alterations and cell type fractions in oncoprints on the Results View (Example)
- Explore the single cell data further in Vitessce on the Patient View (Example)
- Create cohorts and groups based on cell type fractions on the Study View (Example)
- Compare differences in cell type fractions between groups on the Comparison Page (Example)

#
Apr 20, 2022
Added data consisting of 2,557 samples from 5 studies:
- Breast Cancer (HTAN OHSU, 2022) 5 samples
- Colorectal Cancer (MSK, 2022) 47 samples
- Pediatric Pancan Tumors (MSK, 2022) 135 samples
- Sarcoma (MSK, 2022) 2,138 samples
- Lung Cancer in Never Smokers (NCI, Nature Genetics 2021) 232 samples
Gene Tables Update Updated tables of genes (main and alias), based on Jan 1, 2022 HGNC release. See seedDB release note here for details.
Data Improvement
Pan-can studies timeline addition:
TREATMENT,OTHER MALIGNANCY FORM,SAMPLE ACQUISITION,STATUSare added to all 32 TCGA Pan-Can studies. Details for data source and transformation process can be found here or in the README.md files included in each study folder on datahub. Example: patient view of TCGA-A2-A04P in Breast Invasive Carcinoma Tumor Type
Pan-can studies methylation addition: methylation profile (27k and 450k merged) are added to all 32 TCGA Pan-Can studies, in generic assay format. Data source: GDC. Example: search by gene or probe from dropdown, to add a chart in study view, a track in Oncoprint (single study query only), or plots in plots tab.



Single cell (type fraction and phases) data (in generic assay format) is added to Glioblastoma (CPTAC, Cell 2021)
New Feature For the new HTAN OHSU study there is now also an integration with Harvard LSP's Minerva for exploring multiplex imaging:
#
Mar 1, 2022
- New Documentation:
- Add a User Guide
- Add Study View and Query How-to videos
#
Feb 8, 2022
New Feature: Create X vs Y violin plots in Study View using any categorical and numerical clinical data:
#
Jan 19, 2022
New Feature: Numerical filters on the Study View are now editable:
New Feature: In the annotation column choose between showing a single icon OncoKB icon or multiple (one for therapeutic, diagnostic and prognostic):
#
Jan 10, 2022
- New Documentation for Local cBioPortal Installations:
- Explain how to use Genome Nexus to annotate MAF files
#
Jan 4, 2022
Added data consisting of 27,447 samples from 10 studies:
- Endometrial Carcinoma (CPTAC, Cell 2020) 95 samples
- Pancreatic Ductal Adenocarcinoma (CPTAC, Cell 2021) 140 samples
- Lung Squamous Cell Carcinoma (CPTAC, Cell 2021) 108 samples
- Lung Adenocarcinoma (CPTAC, Cell 2020) 110 samples
- Glioblastoma (CPTAC, Cell 2021) 99 samples
- Breast Cancer (CPTAC, Cell 2020) 122 samples
- Pediatric Brain Cancer (CPTAC/CHOP, Cell 2020) 218 samples
- Metastatic Prostate Cancer (Provisional, June 2021) 123 samples
- MSK MetTropism (MSK, Cell 2021) 25,775 samples
- Cancer Therapy and Clonal Hematopoiesis (MSK, 2021) 657 samples
Added TMB (nonsynonymous) scores for all studies. Example: new TMB field for study gbm_cptac_2021
(Details for the calculation can be found HERE)

#
Nov 12, 2021
- Added data consisting of 3,680 samples from 6 studies:
- Breast Cancer MAPK (MSKCC, Nat Commun 2021) 145 samples
- Colorectal Cancer (MSK, 2020) 64 samples
- Breast Cancer (MSK, Clinical Cancer Res 2020) 60 samples
- High-Grade Serous Ovarian Cancer (MSK, 2021) 45 samples
- Diffuse Glioma (GLASS Consortium, Nature 2019) 444 samples
- Pan-cancer analysis of whole genomes (ICGC/TCGA, Nature 2020) 2,922 samples
#
Nov 3,2021
New Feature: Add Uniprot topology as a new annotation track on the Mutations Tab of the Results View. Example: EGFR in MSK-IMPACT (2017) cohort

#
Oct 1, 2021
New Feature: Arm level Copy Number events are now loaded into cBioPortal using the Categorial Generic Assay Data Type. They can be found in a tab under the Add Charts Button of the Study View Example: Arm Level Data in TCGA PanCancer Atlas

#
Sep 22, 2021
- Added data consisting of 14,844 samples from 7 studies:
- Colorectal Cancer (MSK, Gastroenterology 2020) 471 samples
- Metastatic Breast Cancer (MSK, Cancer Discovery 2021) 1,365 samples
- Lung Adenocarcinoma (MSKCC, 2021) 186 samples
- Race Differences in Prostate Cancer (MSK, 2021) 2,069 samples
- Medulloblastoma (DKFZ, Nature 2017) 491 samples
- Thoracic Cancer (MSK, 2021) 68 samples
- China Pan-cancer (OrigiMed, 2020) 10,194 samples
#
Sep 21, 2021
Enhancement: Dowloading the Lollipop plot on the Mutations Tab of the Results View will now also include the annotation tracks:

#
Aug 17, 2021
New Feature: The Mutations Tab of the Results View can now show exon numbers as an annotation track Example: MET Exon 14 Mutations in MSK-IMPACT (2017) cohort

#
Aug 10, 2021
New Feature: Use the filtering capabilities in the Mutations Tab of the Results View to create a custom cohort that one can open directly in the Study View Example: CTNNB1 in MSK-IMPACT (2017) cohort

#
Jul 27, 2021
New Feature: Add a custom filter to any column of the Mutations Tab in the Results View Example: CTNNB1 in MSK-IMPACT (2017) cohort

New Feature: Show detailed descriptions for each annotation source in the header of the the Mutations Table in both the Results View and the Patient View Example link

#
Jul 6, 2021
New Feature: Add any clinical data as a column on the Mutations Tab in the Results View Example: EGFR in MSK-IMPACT (2017) cohort

#
June 23, 2021
- Added data consisting of 1,084 samples from 5 studies:
- Intrahepatic Cholangiocarcinoma (MSK, Hepatology 2021) 412 samples
- Intrahepatic Cholangiocarcinoma (Mount Sinai 2015) 8 samples
- RAD51B Associated Mixed Cancers (Mandelker 2021 19 samples
- Intrahepatic Cholangiocarcinoma (MSK, 2020) 219 samples
- Lung Adenocarcinoma (NPJ Precision Oncology, MSK 2021) 426 samples
- Added mass-spec proteome data from CPTAC to Breast Invasive Carcinoma (TCGA, PanCancer Atlas), Ovarian Serous Cystadenocarcinoma (TCGA, PanCancer Atlas) and Colorectal Adenocarcinoma (TCGA, PanCancer Atlas).
- Added mass-spec phosphoproteome site level expression from CPTAC to Breast Invasive Carcinoma (TCGA, PanCancer Atlas) and Ovarian Serous Cystadenocarcinoma (TCGA, PanCancer Atlas).
- Updated gene tables Updated tables of genes (main and alias), based on HGNC. See details HERE in section
Contents of seed database. Sripts/resources/process used to construct new tables are described HERE.
#
June 1, 2021
New Feature: In certain studies where we have the data we show read counts for uncalled mutations on the Patient View Example: A patient in the Glioma (MSK, 2019) cohort

#
May 10, 2021
New Feature: Pick color for User Defined Groups Example: Color Bladder Cancer Group in MSK-IMPACT (2017) cohort, implemented by The Hyve.

#
May 4, 2021
New Feature: Add more categories of mutations to the Mutations Tab on the Results View, including Driver/VUS, Splice and Structural Variants Example: TP53 alterations in the MSK-IMPACT (2017) cohort

#
April 21, 2021
- Added data consisting of 4074 samples from 9 studies:
- Metaplastic Breast Cancer (MSK, 2021) 19 samples
- Lung Adenocarcinoma (MSKCC, 2020) 604 samples
- Cutaneous Squamous Cell Carcinoma (UCSF, 2021) 105 samples
- MSK-IMPACT and MSK-ACCESS Mixed Cohort (MSK, 2021) 1446 samples
- Melanoma (MSKCC, 2018) 720 samples
- Cholangiocarcinoma (ICGC, Cancer Discov 2017) 489 samples
- Esophageal/Stomach Cancer (MSK, 2020) 487 samples
- Retinoblastoma (MSK, Cancers 2021) 83 samples
- Combined Hepatocellular and Intrahepatic Cholangiocarcinoma (Peking University, Cancer Cell 2019) 121 samples
#
April 20, 2021
New Feature: Add driver annotations to download tab on Results View Example: RAS/RAF alterations in colorectal cancer

#
March 30, 2021
Enhancement: Add 95% Confidence Interval for Survival Plots Example: Altered vs Unaltered EGFR in Lung Cancer

#
March 11, 2021
New Feature: Combine different types of alterations in Comparison View Example: Deletions and Truncating events in primary vs metastases or read more on The Hyve's blog

Enhancement: Improve UI for OncoPrint, aggregating various data modalities in a single add track dropdown button Example: Add clinical, heatmap and treatment response data into the OncoPrint

#
February 16, 2021
Enhancement: Show only TCGA PanCancer Atlas Pathways in Results and Patient View to avoid showing many similar pathways Example: Clinvar APC and CTNNB1 alterations in WNT pathway

#
January 28, 2021
New Feature: Show ClinVar Interpretation in Mutation tables Example: Clinvar Interpretations in BRCA2

#
January 12, 2021
New Feature: Add your own custom data for a sample or patient to use on the study or comparison view Example: Add custom data to three samples and do a comparison

New Feature: Show the mutations of a patient inside a pathway schematic using PathwayMapper Example: Notch signaling pathway in a prostate cancer patient

New Feature: Display and compare generic assays, such as microbiome and treatment response, on the study view Example: Prasinovirus microbiome signatures in TCGA

New Feature: The Plots tab on Results View now allows you to group alterations by Driver and VUS Example: POLE driver mutations vs VUSs against mutation counts in TCGA Colorectal Adenocarcinoma

#
December 31, 2020
- Added data consisting of 430 samples from 5 studies:
- Juvenile Papillomatosis and Breast Cancer (MSK, 2020) 5 samples
- Mixed cfDNA (MSKCC, 2020) 229 samples
- Metastatic Melanoma (DFCI, Nature Medicine 2019) 144 samples
- Lung Cancer (SMC, Cancer Research 2016) 22 samples
- Upper Tract Urothelial Carcinoma (IGBMC, Genome Biology 2021) 30 samples
- Added survival data to Breast Cancer (METABRIC, Nature 2012 & Nat Commun 2016)
#
November 3, 2020
New Feature: The map of local installations of cBioPortal is available now. Please consider registering your instance here.

Enhancement: upgraded the Genomic Evolution tab in Patient View with timeline Example

#
October 20, 2020
- Enhancement: Expression tab has now been merged into the Plots tab

#
October 16, 2020
Added data consisting of 25,078 samples from 5 studies:
- Melanomas (TCGA, Cell 2015) 359 samples
- Retinoblastoma cfDNA (MSKCC 2020) 14 samples
- The Angiosarcoma Project (Provisional, July 2020) 83 samples
- Bladder Cancer (MSK/TCGA, 2020) 476 samples
- Cancer Therapy and Clonal Hematopoiesis (MSK, 2020) 24,146 samples
Added MSI data (MSIsensor from Mariamidze et al. 2018 and MANTIS scores from Roychowdhury et al. 2017) for all 32 TCGA PanCan Atlas Cohorts.

Added new profile “RNA-Seq V2 expression Z-scores relative to normal samples” for 16 TCGA PanCan Atlas Cohorts. The normals samples RNA-Seq V2 expression data were curated from GDC, and can be downloaded from our Datahub or Data Set page. Example: ERBB2 expression z-scores relative to normal expression

#
October 13, 2020
Enhancement: Study View now allows comparing samples with mutations or copy number alterations in different genes

New Feature: When treatment timeline is available (e.g. in this study), Study View now allows the selection and comparison of patients treated with specific drugs, or samples sequenced pre or post specific drug treatments

#
September 30, 2020
- New Feature: Microbiome signature data is available for comparison now. Example: comparing colorectal subtypes for enriched microbiome signatures

#
September 22, 2020
Enhancement: The timeline feature in Patient View has been refactored with an improved UI. Example

Enhancement: Logrank p-values are now provided for all survival analysis (previously only availalbe when comparing two groups). Example

#
August 11, 2020
New Feature: microbiome data of TCGA samples from Poore et al. 2020 are now available for analysis in the OncoPrint and Plots tabs. Example: Orthohepadnavirus across TCGA cancers

New Feature: You can now compare DNA Methylation data between groups using the Comparison feature. Example: Comparing DNA methylation levels between samples with high vs low BRCA1 expression

Added data consisting of 513 samples from 3 studies:
- Breast Cancer (SMC 2018) 187 samples
- Germ Cell Tumors and Shared Leukemias (MSK 2020) 21 samples
- Lung Adenocarcinoma (OncoSG, Nat Genet 2020) 305 samples
Added RPPA data in addition to the microbiome data for 31 TCGA Pancan studies (except LAML)

#
July 21, 2020
New Feature: The Mutations tab now has the option to show mutation effects for different transcripts / isoforms. Note that some annotation features are only available for the canonical isoform. example

Enhancement: The Plots tab is now supported in multi-study queries. example

New Feature: You can now share custom groups in the Study View example

#
June 11, 2020
- Added data consisting of 267 samples from 2 studies:
- Gastric Cancer (OncoSG, 2018) 147 samples
- 120 ctDNA samples added to Non-Small Cell Lung Cancer (TRACERx, NEJM & Nature 2017) 447 samples
#
June 9, 2020
Enhancement: using OQL to query for mutations based on a protein position range. example

New Feature: you can now send the OncoPrint data to the OncoPrinter tool for customization.

Enhancement: Mutational spectrum data can be downloaded from OncoPrint

#
June 2, 2020
- Enhancement: Pediatric cancer studies are now grouped and highlighted in the query page

#
May 6, 2020
- Added data consisting of 574 samples from 3 studies:
- Updated one study:
- Expression data was added to The Metastatic Breast Cancer Project (Provisional, February 2020).
#
April 24, 2020
New Feature: Add a new chart on the Study View for selecting samples based on pre-defined case lists:

#
April 10, 2020
New Feature: Make cohorts on the Study View using continuous molecular profiles of one or more gene(s), such as mRNA expression, methylation, RPPA and continuous CNA. example

Combine this with the group comparison feature to compare e.g. all quartiles of expression:

New Feature: Annotate mutations using the Mutation Mapper Tool on the GRCh38 reference genome:
#
April 3, 2020
- New Feature: Extended the Comparison tab to support the comparison of altered samples per gene or alteration. This example query compares NSCLC patients with 1) both mutated and amplified EGFR, 2) mutated EGFR only, and 3) amplified EGFR only.

#
March 27, 2020
Enhancement: User selections in the Plots tab are now saved in the URL. example
New Feature: Added table of data availability per profile in the Study View. example

#
March 20, 2020
- Enhancement: Extended Survival Analysis to support more outcome measures. example

#
March 18, 2020
- Added data consisting of 1,393 samples from 3 studies:
- Breast Cancer (Alpelisib plus AI, Nature Cancer 2020) 141 samples
- Glioma (MSKCC, Clin Cancer Res 2019) 1,004 samples
- Mixed cfDNA (MSK, Nature Medicine 2019) 248 samples
#
March 3, 2020
New Feature: Added Pathways tab to the Results View page, which visualizes the alteration frequencies of genes in pathways of interest. The pathways are pulled from https://www.pathwaymapper.org and shown in a read only view. One can edit these pathways in the PathwayMapper editor. For more information see the tutorial.
#
February 12, 2020
- Added data consisting of 1,605 samples from 3 studies:
- Tumors with TRK fusions (MSK, 2019) 106 samples
- Lymphoma Cell Lines (MSKCC, 2020) 34 samples
- Prostate Adenocarcinoma (MSKCC, 2020) 1,465 samples
#
February 6, 2020
New Feature: Extend the recent group comparison feature by allowing comparisons inside the Results View page. The new tab allows for quick comparison of altered vs unaltered cases by survival, clinical information, mutation, copy number events and mRNA expression:
Performance enhancement: the Study View's mutation table now loads faster for studies with multiple gene panels. For the genie portal, which has a study with many different gene panels this resulted in a speed-up from ~90-120 seconds to 5 seconds.
Read more about the v3.2.2 release here
#
January 30, 2020
Enhancement: Show HGVSg in mutations table and linkout to Genome Nexus:

Enhancement: Add a pencil button near gene list in results page which opens interface for quickly modifying the oql of the query:

See more updates here
#
January 29, 2020
- Added data consisting of 197 samples from 2 studies:
- Bladder/Urinary Tract Cancer (MSK, 2019) 78 samples
- Upper Tract Urothelial Carcinoma (MSK, 2019) 119 samples
#
December 19, 2019
Enhancement: We restored support for submitting large queries from external applications using HTTP POST requests. Accepted parameters are the same as appear in the url of a query submitted from the homepage.
See more updates here
#
December 12, 2019
Enhancement: Several enhancements to the display of gene panels on the Patient View page, by The Hyve, described in more detail here
Enhancement: Add Count Bubbles to Oncoprint Toolbar

See more updates here
#
November 29, 2019
- Enhancement: Support group comparison for custom charts in Study View page
- Enhancement: Performance improvement of Co-Expression analysis.
- Enhancement: Kaplan-Meier plots now supports custom time range.
- See more updates here

#
November 22, 2019
- New Feature: Support for Treatment response data in the Oncoprint and Plots tab, including new Waterfall plot type. Read more in The Hyve's blog post
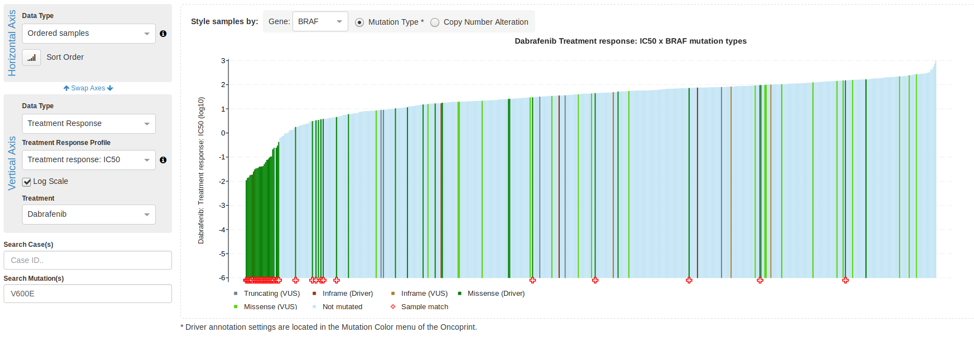
#
November 15, 2019
- Enhancement: heatmap tracks in OncoPrint now has separate headers and sub-menus. example

- Enhancement: global settings for query session

#
November 7, 2019
- Added data consisting of 212 samples from 3 studies:
- Metastatic Melanoma (DFCI, Science 2015) 110 samples
- Melanoma (MSKCC, NEJM 2014) 64 samples
- Metastatic Melanoma (UCLA, Cell 2016) 38 samples
#
October 30, 2019
- Added data consisting of 178 samples from 2 studies:
#
October 23, 2019
- Enhancement: Quick example links in Plots tab. example
#
October 14, 2019
- New Feature: Fusion Genes table in
Study View. example

#
October 11, 2019
Enhancement: The Download interface on the homepage has been removed. Enhanced download functionality is now available after querying on the results page.
Home page:

Results page:

Note that as before one can always download the full raw data on the Data Sets page or from Datahub.
#
October 9, 2019
Added data consisting of 2725 samples from 4 studies:
- Cancer Cell Line Encyclopedia (Broad, 2019) 1739 samples
- Chronic Lymphocytic Leukemia (Broad, Nature 2015) 537 samples
- Rectal Cancer (MSK,Nature Medicine 2019) 339 samples
- Colon Cancer (CPTAC-2 Prospective, Cell 2019) 110 samples
Updated Esophageal Carcinoma (TCGA, Nature 2017) with addition of CNA data for Esophageal Squamous Cell Carcinoma cases 90 samples.
#
September 18, 2019
- New Feature: The list and order of charts of a study will be automatically saved now as a user preference on the study view page.
#
September 6, 2019
- Added data consisting of 1216 samples from 3 studies:
- Breast Cancer (MSKCC, 2019) 70 samples
- Brain Tumor PDXs (Mayo Clinic, 2019)97 samples
- Adenoid Cystic Carcinoma Project (2019) 1049 samples
#
August 13, 2019
- Added data consisting of 295 samples from 3 studies:
- Pediatric Preclinical Testing Consortium (PPTC, 2019) 261 samples
- Non-small cell lung cancer (MSK, Science 2015) 16 samples
- Prostate Cancer (MSK, 2019) 18 samples
#
July 26, 2019
- Added data consisting of 35 samples from 1 study:
- Added Hypoxia data for:
- Breast Invasive Carcinoma (TCGA, PanCancer Atlas)
- Colorectal Adenocarcinoma (TCGA, PanCancer Atlas)
- Cervical Squamous Cell Carcinoma (TCGA, PanCancer Atlas)
- Glioblastoma Multiforme (TCGA, PanCancer Atlas)
- Head and Neck Squamous Cell Carcinoma (TCGA, PanCancer Atlas)
- Kidney Renal Clear Cell Carcinoma (TCGA, PanCancer Atlas)
- Kidney Renal Papillary Cell Carcinoma (TCGA, PanCancer Atlas)
- Brain Lower Grade Glioma (TCGA, PanCancer Atlas)
- Liver Hepatocellular Carcinoma (TCGA, PanCancer Atlas)
- Lung Adenocarcinoma (TCGA, PanCancer Atlas)
- Lung Squamous Cell Carcinoma (TCGA, PanCancer Atlas)
- Ovarian Serous Cystadenocarcinoma (TCGA, PanCancer Atlas)
- Pancreatic Adenocarcinoma (TCGA, PanCancer Atlas)
- Pheochromocytoma and Paraganglioma (TCGA, PanCancer Atlas)
- Prostate Adenocarcinoma (TCGA, PanCancer Atlas)
- Skin Cutaneous Melanoma (TCGA, PanCancer Atlas)
- Thyroid Carcinoma (TCGA, PanCancer Atlas)
- Uterine Corpus Endometrial Carcinoma (TCGA, PanCancer Atlas)
#
July 24, 2019
- Added data consisting of 151 samples from 1 study:
- Myeloproliferative Neoplasms (CIMR, NEJM 2013) 151 samples
#
July 13, 2019
- Public Release 6.1 of AACR Project GENIE:
- The sixth data set, GENIE 6.0-public, was released in early July 2019. A patch to GENIE 6.0-public, GENIE 6.1-pubic, was subsequently released on July 13, 2019. The combined data set now includes nearly 70,000 de-identified genomic records collected from patients who were treated at each of the consortium's participating institutions, making it among the largest fully public cancer genomic data sets released to date. The combined data set now includes data for nearly 80 major cancer types, including data from nearly 11,000 patients with lung cancer, greater than 9,700 patients with breast cancer, and nearly 7,000 patients with colorectal cancer.
- More detailed information can be found in the AACR GENIE Data Guide. In addition to accessing the data via the cBioPortal, users can download the data directly from Sage Bionetworks. Users will need to create an account for either site and agree to the terms of access.
- For frequently asked questions, visit the AACR FAQ page.
#
July 2, 2019
- Added data consistng of 785 samples from 4 studies:
- Non-Small Cell Lung Cancer (TRACERx, NEJM 2017) 327 samples
- Acute myeloid leukemia or myelodysplastic syndromes (WashU, 2016) 136 samples
- Basal Cell Carcinoma (UNIGE, Nat Genet 2016) 293 samples
- Colon Adenocarcinoma (CaseCCC, PNAS 2015) 29 samples
#
June 19, 2019
New Feature: Show Genome Aggregation Database (gnomAD) population frequencies in the mutations table - see example:

#
June 12, 2019
- Added data of 1350 samples from 3 studies:
- Pheochromocytoma and Paraganglioma (TCGA, Cell 2017) 178 samples
- Metastatic Solid Cancers (UMich, Nature 2017) 500 samples
- Acute Myeloid Leukemia (OHSU, Nature 2018) 672 samples
- Added survival data for TCGA PanCan Atlas Cohorts (>10,000 samples across 33 tumor types).
- Added hypoxia data for Bladder Urothelial Carcinoma (TCGA, PanCancer Atlas)
#
June 7, 2019
- New Group Comparison Feature: Compare clinical and genomic features of user-defined groups of samples/patients. View Tutorial
#
May 8, 2019
New Feature: Show Post Translational Modification (PTM) information from dbPTM on the Mutation Mapper - see example:

#
April 26, 2019
- Added data of 568 samples from 4 studies:
#
March 29, 2019
New Feature: Use the new quick search tab on the homepage to more easily navigate to a study, gene or patient:

#
March 15, 2019
- Added data of 338 samples from 4 studies:
- Adenoid Cystic Carcinoma (MGH, Nat Gen 2016) 10 samples
- Gallbladder Cancer (MSK, Cancer 2018) 103 samples
- The Metastatic Prostate Cancer Project (Provisional, December 2018) 19 samples
- Adult Soft Tissue Sarcomas (TCGA, Cell 2017) 206 samples
#
February 22, 2019
- Enhancement: Exon number and HGVSc annotations are available in optional columns in the Mutations tab on the Results page and in the Patient View.
- New feature: option to a show regression line in the scatter plot in the Plots tab on the Results page

#
February 19, 2019
- New feature: Copy-Number Segments tab on the Study View page using igv.js v2 - see example
- Improved Copy-Number Segments tab on the Results page
- New feature: OncoKB and Cancer Hotspots tracks in the Mutations tab on the Results page

#
January 24, 2019
- Added data of 2328 samples from 8 studies:
- Uveal Melanoma (QIMR, Oncotarget 2016) 28 samples
- Squamous Cell Carcinoma of the Vulva (CUK, Exp Mol Med 2018) 15 samples
- TMB and Immunotherapy (MSKCC, Nat Genet 2019) 1661 samples
- Glioma (MSK, 2018) 91 samples
- Urothelial Carcinoma (Cornell/Trento, Nat Gen 2016) 72 samples
- Hepatocellular Carcinoma (MSK, Clin Cancer Res 2018) 127 samples
- MSK Thoracic PDX (MSK, Provisional) 139 samples
- Cholangiocarcinoma (MSK, Clin Cancer Res 2018) 195 samples
- Updated data for The Metastatic Breast Cancer Project (Provisional, October 2018) 237 samples
#
January 10, 2019
- cBioPortal now supports queries for driver mutations, fusions and copy number alterations as well as germline/somatic mutations using Onco Query Language (OQL) -- see example
- A new tutorial explores OQL and provides examples of how OQL can be a powerful tool to refine queries.
#
December 17, 2018
- The 10th phase of cBioPortal architectural upgrade is now complete: the Study View has been moved to the new architecture with numerous improvements. This marks the completion of the cBioPortal architectural refactoring! 🎉🎉🎉
#
October 29, 2018
- The ninth phase of the cBioPortal architectural upgrade is now complete: the results page is now a single-page application with better performance.
- Supported plotting mutations by type in Plots tab

#
October 19, 2018
- Support selection of transcript of interest in the MutationMapper tool via Genome Nexus.

#
October 17, 2018
- Added data of 3578 samples from 8 studies:
- Rhabdoid Cancer (BCGSC, Cancer Cell 2016) 40 samples
- Diffuse Large B-Cell Lymphoma (Duke, Cell 2017) 1001 samples
- Diffuse Large B cell Lymphoma (DFCI, Nat Med 2018) 135 samples
- Breast Fibroepithelial Tumors (Duke-NUS, Nat Genet 2015) 22 samples
- Uterine Clear Cell Carcinoma (NIH, Cancer 2017) 16 samples
- Endometrial Cancer (MSK, 2018) 197 samples
- Breast Cancer (MSK, Cancer Cell 2018) 1918 samples
- MSS Mixed Solid Tumors (Van Allen, 2018) 249 samples
- Updated data for The Angiosarcoma Project (Provisional, September 2018) 48 samples
#
August 20, 2018
- Now you can log in on the public cBioPortal with your Google account and save your virtual studies for quick analysis.

#
August 7, 2018
- The eighth phase of the cBioPortal architectural upgrade is now complete: The Plots, Expression, Network, and Bookmarks tabs, and therefore all analysis tabs in the results page, have been moved to the new architecture.
- Updated the MutationMapper tool, now connecting to Genome Nexus for annotating mutations on the fly.
- Total Mutations and Fraction Genome Altered are now available in Plots tab for visualization and analysis.
- Enhanced clinical attribute selector for OncoPrint, now showing sample counts per attribute.

#
July 27, 2018
- Added data of 2787 samples from 10 studies:
- Mixed Tumors (PIP-Seq 2017) 103 samples
- Nonmuscle Invasive Bladder Cancer (MSK Eur Urol 2017) 105 samples
- Pediatric Neuroblastoma (TARGET, 2018) 1089 samples
- Pediatric Pan-Cancer (DKFZ - German Cancer Consortium, 2017) 961 samples
- Skin Cutaneous Melanoma (Broad, Cancer Discov 2014) 78 samples
- Cutaneous Squamous Cell Carcinoma (MD Anderson, Clin Cancer Res 2014) 39 samples
- Diffuse Large B-cell Lymphoma (BCGSC, Blood 2013) 53 samples
- Non-Hodgkin Lymphoma (BCGSC, Nature 2011) 14 samples
- Chronic lymphocytic leukemia (ICGA, Nat 2011) 105 samples
- Neuroblastoma (Broad Institute 2013) 240 samples
#
June 20, 2018
- The seventh phase of the cBioPortal architectural upgrade is now complete: The Enrichments and Co-Expression tabs have been moved to the new architecture.
- Supported merged gene tracks in OncoPrint and Onco Query Language -- see example

#
May 10, 2018
- Enhanced OncoPrint to show germline mutations -- see example

#
April 17, 2018
- Added data of 3732 samples from 4 TARGET studies:
- Pediatric Acute Lymphoid Leukemia - Phase II (TARGET, 2018) 1978 samples
- Pediatric Acute Myeloid Leukemia (TARGET, 2018) 1025 samples
- Pediatric Rhabdoid Tumor (TARGET, 2018) 72 samples
- Pediatric Wilms' Tumor (TARGET, 2018) 657 samples
- Added data of 3416 samples from 10 published studies:
- Prostate Adenocarcinoma (MSKCC/DFCI, Nature Genetics 2018) 1013 samples
- Prostate Adenocarcinoma (EurUrol, 2017) 65 samples
- Non-Small Cell Lung Cancer (MSK, JCO 2018) 240 samples
- Small-Cell Lung Cancer (Multi-Institute 2017) 20 samples
- The Angiosarcoma Project (Provisional, February 2018) 14 samples
- Acute Lymphoblastic Leukemia (St Jude, Nat Genet 2016) 73 samples
- Updated Segment data and Allele Frequencies for The Metastatic Breast Cancer Project (Provisional, October 2017) 103 samples
- Colorectal Cancer (MSK, Cancer Cell 2018) 1134 samples
- Metastatic Esophagogastric Cancer (MSK,Cancer Discovery 2017) 341 samples
- Bladder Cancer (TCGA, Cell 2017) 413 samples
#
April 5, 2018
- Added data from the TCGA PanCanAtlas project with >10,000 samples from 33 tumor types:
- Adrenocortical Carcinoma (TCGA, PanCancer Atlas)
- Bladder Urothelial Carcinoma (TCGA, PanCancer Atlas)
- Breast Invasive Carcinoma (TCGA, PanCancer Atlas)
- Cervical Squamous Cell Carcinoma (TCGA, PanCancer Atlas)
- Colon Adenocarcinoma (TCGA, PanCancer Atlas)
- Cholangiocarcinoma (TCGA, PanCancer Atlas)
- Diffuse Large B-Cell Lymphoma (TCGA, PanCancer Atlas)
- Esophageal Adenocarcinoma (TCGA, PanCancer Atlas)
- Glioblastoma Multiforme (TCGA, PanCancer Atlas)
- Head and Neck Squamous Cell Carcinoma (TCGA, PanCancer Atlas)
- Kidney Chromophobe (TCGA, PanCancer Atlas)
- Kidney Renal Clear Cell Carcinoma (TCGA, PanCancer Atlas)
- Kidney Renal Papillary Cell Carcinoma (TCGA, PanCancer Atlas)
- Acute Myeloid Leukemia (TCGA, PanCancer Atlas)
- Brain Lower Grade Glioma (TCGA, PanCancer Atlas)
- Liver Hepatocellular Carcinoma (TCGA, PanCancer Atlas)
- Lung Adenocarcinoma (TCGA, PanCancer Atlas)
- Lung Squamous Cell Carcinoma (TCGA, PanCancer Atlas)
- Mesothelioma (TCGA, PanCancer Atlas)
- Ovarian Serous Cystadenocarcinoma (TCGA, PanCancer Atlas)
- Pancreatic Adenocarcinoma (TCGA, PanCancer Atlas)
- Pheochromocytoma and Paraganglioma (TCGA, PanCancer Atlas)
- Prostate Adenocarcinoma (TCGA, PanCancer Atlas)
- Rectum Adenocarcinoma (TCGA, PanCancer Atlas)
- Sarcoma (TCGA, PanCancer Atlas)
- Skin Cutaneous Melanoma (TCGA, PanCancer Atlas)
- Stomach Adenocarcinoma (TCGA, PanCancer Atlas)
- Testicular Germ Cell Tumors (TCGA, PanCancer Atlas)
- Thyroid Carcinoma (TCGA, PanCancer Atlas)
- Thymoma (TCGA, PanCancer Atlas)
- Uterine Corpus Endometrial Carcinoma (TCGA, PanCancer Atlas)
- Uterine Carcinosarcoma (TCGA, PanCancer Atlas)
- Uveal Melanoma (TCGA, PanCancer Atlas)
#
March 20, 2018
- The sixth phase of the cBioPortal architectural upgrade is now complete: The Download tab has been moved to the new architecture.
- Data can now be downloaded in tabular format from OncoPrint.
- Added an option to download an SVG file on the Cancer Type Summary tab.
#
January 15, 2018
- The fifth phase of the cBioPortal architectural upgrade is now complete: The OncoPrint and Survival tabs have been moved to the new architecture.
#
November 20, 2017
- You can now combine multiple studies and view them on the study summary page. Example: liver cancer studies
- You can now bookmark or share your selected samples as virtual studies with the share icon on the study summary page. Example: a virtual study of breast tumors
- Cross-study query reimplemented: Now you can view an OncoPrint of multiple studies. Example: querying NSCLC tumors from 5 studies

#
October 17, 2017
- The fourth phase of the cBioPortal architectural upgrade is now complete: The Mutual Exclusivity and Cancer Type Summary tabs have been moved to the new architecture.
- Updated protein structure alignment data in Mutations tab are now retrieved from Genome Nexus via the G2S web service.
#
October 2, 2017
- Added data of 1646 samples from 7 published studies:
- NGS in Anaplastic Oligodendroglioma and Anaplastic Oligoastrocytomas tumors (MSK, Neuro Oncol 2017) 22 samples
- MSK-IMPACT Clinical Sequencing Cohort for Non-Small Cell Cancer (MSK, Cancer Discovery 2017) 915 samples
- Paired-exome sequencing of acral melanoma (TGEN, Genome Res 2017) 38 samples
- MSK-IMPACT Clinical Sequencing Cohort in Prostate Cancer (MSK, JCO Precision Oncology 2017) 504 samples
- Whole-exome sequences (WES) of pretreatment melanoma tumors (UCLA, Cell 2016) 39 samples
- Next generation sequencing (NGS) of pre-treatment metastatic melanoma samples (MSK, JCO Precision Oncology 2017) 66 samples
- Targeted gene sequencing in 62 high-grade primary Unclassified Renal Cell Carcinoma (MSK, Nature 2016) 62 samples
- Updated data for MSK-IMPACT Clinical Sequencing Cohort (MSK, Nat Med 2017) with overall survival data.
#
August 3, 2017
- The third phase of the cBioPortal architectural upgrade is now complete: The Mutations tab now has a fresh look and faster performance -- see example

- Variant interpretations from the CIViC database are now integrated into the annotation columns on the Mutations tab and in the patient view pages
- New summary graph for all cancer studies and samples on the front page
#
June 26, 2017
- The second phase of the cBioPortal architectural upgrade is now complete: The query interface now has a fresh look and faster performance.

#
May 12, 2017
- Added data of 12,211 samples from 11 published studies:
- MSK-IMPACT Clinical Sequencing Cohort (MSK, Nat Med 2017) 10,945 samples
- Whole-genome sequencing of pilocytic astrocytomasatic (DKFZ, Nat Genetics, 2013) 96 samples
- Hepatocellular Carcinomas (INSERM, Nat Genet 2015) 243 samples
- Cystic Tumor of the Pancreas (Johns Hopkins, PNAS 2011) 32 samples
- Whole-Genome Sequencing of Pancreatic Neuroendocrine Tumors (ARC- Net, Nature, 2017) 98 samples
- Medulloblastoma (Sickkids, Nature 2016) 46 samples
- Genetic Characterization of NSCLC young adult patients (University of Turin, Lung Cancer 2016) 41 samples
- Genomic Profile of Patients with Advanced Germ Cell Tumors (MSK, JCO 2016). 180 samples
- Ampullary Carcinoma (Baylor, Cell Reports 2016) 160 samples
- Mutational profiles of metastatic breast cancer (INSERM, 2016) 216 samples
- Prostate Adenocarcinoma (Fred Hutchinson CRC, Nat Med 2016) 154 samples
#
May 5, 2017
- First phase of cBioPortal architectural upgrade complete: Patient view now has fresh look and faster performance. example
#
March 28, 2017
New features:
- Per-sample mutation spectra are now available in OncoPrints -- see example

- mRNA heat map clustering is now supported in OncoPrints
- MDACC Next-Generation Clustered Heat Maps are now available in the patient view
- cBioPortal web site style change
#
Feburary 2, 2017
- New features:
- 3D hotspot mutation annotations are now available from 3dhotspots.org
- New data:
- CPTAC proteomics data have been integrated for TCGA breast, ovarian, and colorectal provisional studies
#
December 23, 2016
New features:
- Heat map visualization of gene expression data in the OncoPrint

- Heat map visualization of gene expression data in the Study View page connecting to MDACC's TCGA Next-Generation Clustered Heat Map Compendium
#
October 7, 2016
New features:
- All data sets can now be downloaded as flat files from the new Data Hub
- Annotation of putative driver missense mutations in OncoPrints, based on OncoKB, mutation hotspots, and recurrence in cBioPortal and COSMIC

- Copy number segments visualization directly in the browser in a new CN Segments tab via IGV.js

Improvements:
- Improved cancer study view page (bug fixes and increased performance)
#
July 24, 2016
- Added data of 4,375 samples from 21 published studies:
- Adenoid Cystic Carcinoma (MDA, Clin Cancer Res 2015) 102 samples
- Adenoid Cystic Carcinoma (FMI, Am J Surg Pathl. 2014) 28 samples
- Adenoid Cystic Carcinoma (Sanger/MDA, JCI 2013) 24 samples
- Adenoid Cystic Carcinoma of the Breast (MSKCC, J Pathol. 2015) 12 samples
- Bladder Cancer, Plasmacytoid Variant (MSKCC, Nat Genet 2016) 34 samples
- Breast Cancer (METABRIC, Nat Commun 2016) 1980 samples
- Chronic Lymphocytic Leukemia (Broad, Cell 2013) 160 samples
- Chronic Lymphocytic Leukemia (IUOPA, Nature 2015) 506 samples
- Colorectal Adenocarcinoma (DFCI, Cell Reports 2016) 619 samples
- Cutaneous T Cell Lymphoma (Columbia U, Nat Genet 2015) 42 samples
- Diffuse Large B-Cell Lymphoma (Broad, PNAS 2012) 58 samples
- Hepatocellular Adenoma (Inserm, Cancer Cell 2014) 46 samples
- Hypodiploid Acute Lymphoid Leukemia (St Jude, Nat Genet 2013) 44 samples
- Insulinoma (Shanghai, Nat Commun 2013) 10 samples
- Malignant Pleural Mesothelioma (NYU, Cancer Res 2015) 22 samples
- Mantle Cell Lymphoma (IDIBIPS, PNAS 2013) 29 samples
- Myelodysplasia (Tokyo, Nature 2011) 29 samples
- Neuroblastoma (Broad, Nat Genet 2013) 56 samples
- Oral Squamous Cell Carcinoma (MD Anderson, Cancer Discov 2013) 40 samples
- Pancreatic Adenocarcinoma (QCMG, Nature 2016) 383 samples
- Recurrent and Metastatic Head & Neck Cancer (JAMA Oncology, 2016) 151 samples
- New TCGA study:
- Pan-Lung Cancer (TCGA, Nat Genet 2016) 1144 samples
- Updated TCGA provisional studies
- updated to the Firehose run of January 28, 2016
- RPPA data updated with the latest data from MD Anderson
- OncoTree codes assigned per sample
#
June 6, 2016
- New features:
- Annotation of mutation effect and drug sensitivity on the Mutations tab and the patient view pages (via OncoKB)

- Annotation of mutation effect and drug sensitivity on the Mutations tab and the patient view pages (via OncoKB)
- Improvements:
- Improved OncoPrint visualization using WebGL: faster, more zooming flexibility, visualization of recurrent variants
- Improved Network tab with SBGN view for a single interaction
- Performance improvement of tables in the study view page
- Mutation type summary on the Mutations tab
#
March 31, 2016
- New features:
- Visualization of "Enrichments Analysis" results via volcano plots
- Improved performance of the cross cancer expression view by switching to Plot.ly graphs
- Improvements to the "Clinical Data" tab on the study view page
- More customization options for the cross-cancer histograms
- Performance improvements in the study view and query result tabs
- Added data of 1235 samples from 3 published studies:
#
January 12, 2016
- New features:
- Visualization of multiple samples in a patient
- Visualization of timeline data of a patient (example)

- All TCGA data updated to the latest Firehose run of August 21, 2015
- New TCGA studies:
- Added data of 650 samples from 10 published studies:
- Neuroblastoma (AMC Amsterdam, Nature 2012)
- Clear Cell Renal Cell Carcinoma (U Tokyo, Nat Genet 2013)
- Multiregion Sequencing of Clear Cell Renal Cell Carcinoma (IRC, Nat Genet 2014)
- Bladder Urothelial Carcinoma (Dana Farber & MSKCC, Cancer Discovery 2014)
- Low-Grade Gliomas (UCSF, Science 2014)
- Esophageal Squamous Cell Carcinoma (UCLA, Nat Genet 2014)
- Acinar Cell Carcinoma of the Pancreas (Johns Hopkins, J Pathol 2014)
- Gastric Adenocarcinoma (TMUCIH, PNAS 2015)
- Primary Central Nervous System Lymphoma (Mayo Clinic, Clin Cancer Res 2015)
- Desmoplastic Melanoma (Broad Institute, Nat Genet 2015)
- All mutation data mapped to UniProt canonical isoforms
#
December 23, 2015
- New features:
- Visualization of RNA-seq expression levels across TCGA studies (cross-cancer queries)

- Selection of genes in the study view to initiate queries

- Visualization of RNA-seq expression levels across TCGA studies (cross-cancer queries)
- Improvement:
- 3-D structures in the "Mutations" tab are now rendered by 3Dmol.js (previously JSmol)
- Improved performance by code optimization and compressing large data by gzip
#
December 1, 2015
- New feature: Annotated statistically recurrent hotspots, via new algorithm by Chang et al. 2015

#
November 9, 2015
- New features:
- Links to MyCancerGenome.org for mutations

- Improved display of selection samples on the study view page
- Links to MyCancerGenome.org for mutations
- Improvements:
- "Enrichments" analysis is now run across all genes
- The "Network" tab is now using Cytoscape.js (Adobe Flash is no longer required)
#
October 6, 2015
- New TCGA data:
- Added data of 763 samples from 12 published studies:
- Small Cell Lung Cancer (U Cologne, Nature 2015)
- Uterine Carcinosarcoma (JHU, Nat Commun 2014)
- Microdissected Pancreatic Cancer Whole Exome Sequencing (UTSW, Nat Commun 2015)
- Pancreatic Neuroendocrine Tumors (JHU, Science 2011)
- Renal Non-Clear Cell Carcinoma (Genentech, Nat Genet 2014)
- Infant MLL-Rearranged Acute Lymphoblastic Leukemia (St Jude, Nat Genet 2015)
- Rhabdomyosarcoma (NIH, Cancer Discov 2014)
- Thymic epithelial tumors (NCI, Nat Genet 2014)
- Pediatric Ewing Sarcoma (DFCI, Cancer Discov 2014)
- Ewing Sarcoma (Institut Cuire, Cancer Discov 2014)
- Cutaneous squamous cell carcinoma (DFCI, Clin Cancer Res 2015)
- Gallbladder Carcinoma (Shanghai, Nat Genet 2014)
#
August 21, 2015
- All TCGA data updated to the Firehose run of April 16, 2015.
- New feature: Enrichments Analysis finds alterations that are enriched in either altered or unaltered samples.
- Improvement: improved OncoPrint with better performance.
#
June 3, 2015
- Improvements:
- Allowed downloading data in each chart/table in study summary page.
- Added log-rank test p-values to the survival plots in study summary page.
- Improved visualization of patient clinical data in patient-centric view.
- Added option to merge multiple samples for the same patient in OncoPrint.
#
April 28, 2015
- New features:
- Redesigned query interface to allow selecting multiple cancer studies
- Redesigned Plots tab
#
January 20, 2015
- All TCGA data updated to the Firehose run of October 17, 2014
- COSMIC data updated to V71
- New features:
- Query page: better search functions to find cancer studies
- OncoPrints now support color coding of different mutation types
- OncoPrints now support multiple clinical annotation tracks
- OncoPrinter tool now supports mRNA expression changes

#
January 6, 2015
- New feature: You can now view frequencies of mutations and copy-number alterations in the study view. These tables are updated dynamically when selecting subsets of samples.

#
December 9, 2014
- New TCGA data:
- Added complete and up-to-date clinical data for all TCGA provisional studies
- All TCGA data updated to the Firehose run of July 15, 2014
- New TCGA provisional studies: Esophageal cancer, Pheochromocytoma and Paraganglioma (PCPG)
- New published TCGA studies: Thyroid Cancer and Kidney Chromophobe
- Added data of 172 samples from 4 published studies:
- New features:
- Redesigned Mutual Exclusivity tab
- Added correlation scores for scatter plots on the Plots tab
- Download links to GenomeSpace
#
October 24, 2014
- Added data of 885 samples from 11 published studies:
- Colorectal Adenocarcinoma Triplets (MSKCC, Genome Biology 2014)
- Esophageal Squamous Cell Carcinoma (ICGC, Nature 2014)
- Malignant Peripheral Nerve Sheath Tumor (MSKCC, Nature Genetics 2014)
- Melanoma (Broad/Dana Farber, Nature 2012)
- Nasopharyngeal Carcinoma (National University Singapore, Nature Genetics 2014)
- Prostate Adenocarcinoma CNA study (MSKCC, PNAS 2014)
- Prostate Adenocarcinoma Organoids (MSKCC, Cell 2014)
- Stomach Adenocarcinoma (TCGA, Nature 2014)
- Stomach Adenocarcinoma (Pfizer and University of Hong Kong, Nature Genetics 2014)
- Stomach Adenocarcinoma (University of Hong Kong, Nature Genetics 2011)
- Stomach Adenocarcinoma (University of Tokyo, Nature Genetics 2014)
#
August 8, 2014
- Released two new tools
- Oncoprinter lets you create Oncoprints from your own, custom data
- MutationMapper draws mutation diagrams (lollipop plots) from your custom data
#
May 21, 2014
- All TCGA data updated to the Firehose run of April 16, 2014
#
May 12, 2014
- Improved study summary page including survival analysis based on clinical attributes
e.g. TCGA Endometrial Cancer cohort

#
March 27, 2014
- New features:
- Visualizing of mutations mapped on 3D structures (individual or multiple mutations, directly in the browser)
- Gene expression correlation analysis (find all genes with expression correlation to your query genes)
- The Patient-Centric View now displays mutation frequencies across all cohorts in cBioPortal for each mutation
- The Mutation Details Tab and the Patient-Centric View now display the copy-number status of each mutation

#
March 18, 2014
- All TCGA data updated to the Firehose run of January 15, 2014
- Updated to the latest COSMIC data (v68)
- Added two new provisional TCGA studies:
- Adrenocortical Carcinoma
- Uterine Carcinosarcoma
- Added mutation data of 898 samples from 11 published studies:
- Hepatocellular Carcinoma (RIKEN, Nature Genetics 2012)
- Hepatocellular Carcinoma (AMC, Hepatology in press)
- Medulloblastoma (Broad, Nature 2012)
- Medulloblastoma (ICGC, Nature 2012)
- Medulloblastoma (PCGP, Nature 2012)
- Multiple Myeloma (Broad, Cancer Cell 2014)
- Pancreatic Adenocarcinoma (ICGC, Nature 2012)
- Small Cell Carcinoma of the Ovary (MSKCC, Nature Genetics in press)
- Small Cell Lung Cancer (CLCGP, Nature Genetics 2012)
- Small Cell Lung Cancer (Johns Hopkins, Nature Genetics 2012)
- NCI-60 Cell Lines (NCI, Cancer Res. 2012)
#
December 9, 2013
- Added mutation data of 99 bladder cancer samples (BGI, Nature Genetics 2013)
#
December 6, 2013
- Data sets matching four recently submitted or published TCGA studies are now available
- Glioblastoma (Cell 2013)
- Bladder carcinoma (Nature, in press)
- Head & neck squamous cell carcinoma (submitted)
- Lung adenocarcinoma (submitted)
#
November 8, 2013
- All TCGA data updated to the Firehose run of September 23, 2013.
- Updated to the latest COSMIC data (v67).
- Added mutation data of 792 samples from 9 published cancer studies:
- Esophageal Adenocarcinoma (Broad, Nature Genetics 2013)
- Head and Neck Squamous Cell Carcinoma (Broad, Science 2011)
- Head and Neck Squamous Cell Carcinoma (Johns Hopkins, Science 2011)
- Kidney Renal Clear Cell Carcinoma (BGI, Nature Genetics 2012)
- Prostate Adenocarcinoma, Metastatic (Michigan, Nature 2012)
- Prostate Adenocarcinoma (Broad/Cornell, Nature Genetics 2012)
- Prostate Adenocarcinoma (Broad/Cornell, Cell 2013)
- Skin Cutaneous Melanoma (Yale, Nature Genetics 2012)
- Skin Cutaneous Melanoma (Broad, Cell 2012)
#
October 21, 2013
- Improved interface for survival plots, including information on individual samples via mouse-over
- New fusion glyph in OncoPrints

- Improved cross-cancer query: new alteration frequency histogram (example below - query gene: CDKN2A) and mutation diagram

#
September 9, 2013
- Updated COSMIC data (v66 Release)
- Improved / interactive visualization on the "Protein changes" tab
- Enhanced mutation diagrams: color-coding by mutation time and syncing with table filters
- Addition of DNA cytoband information in the patient view of copy-number changes
- OncoPrints now allow the display of an optional track with clinical annotation (Endometrial cancer example below)

#
July 25, 2013
- Multi-gene correlation plots.
- Variant allele frequency distribution plots for individual tumor samples.
- Tissue images for TCGA samples in the patient view, via Digital Slide Archive. Example.
#
July 16, 2013
- All TCGA data updated to the May Firehose run (May 23, 2013).
- TCGA Pancreatic Cancer study (provisional) added.
#
July 4, 2013
- Improved rendering of mutation diagrams, including ability to download in PDF format.
- Improved home page: Searchable cancer study & gene set selectors, data sets selector.
#
June 17, 2013
- Improved interface for correlation plots, including information on individual samples via mouse-over.
- Gene Details from Biogene are now available in the Network view.
- Added mutation and copy number data from a new adenoid cystic carcinoma study: Ho et al., Nature Genetics 2013.
- Added mutation data from 6 cancer studies.
- Breast Invasive Carcinoma (Shah et al., Nature 2012)
- Breast Invasive Carcinoma (Banerji et al., Nature 2012)
- Breast Invasive Carcinoma (Stephens et al., Nature 2012)
- Lung Adenocarcinoma (Imielinksi et al., Cell 2012)
- Lung Adenocarcinoma (Ding et al., Nature 2008)
- Colorectal Cancer (Seshagiri et al., Nature 2012)
#
June 4, 2013
- All TCGA data updated to the April Firehose run (April 21, 2012).
#
May 14, 2013
- Added a published TCGA study: Acute Myeloid Leukemia (TCGA, NEJM 2013).
#
April 28, 2013
- All TCGA data updated to the March Firehose run (March 26, 2012).
- mRNA percentiles for altered genes shown in patient view.
#
April 2, 2013
- All TCGA data updated to the February Firehose run (February 22, 2012).
#
March 28, 2013
- All TCGA data updated to the January Firehose run (January 16, 2012).
- Data from a new bladder cancer study from MSKCC has been added (97 samples, Iyer et al., JCO in press).
#
February 16, 2013
- The cBio Portal now contains mutation data from all provisional TCGA projects. Please adhere to the TCGA publication guidelines when using these and any TCGA data in your publications.
- All data updated to the October Firehose run (October 24, 2012).
- Sequencing read counts and frequencies are now shown in the Mutation Details table when available.
- Improved OncoPrints, resulting in performance improvements.
#
November 21, 2012
- Major new feature: Users can now visualize genomic alterations and clinical data of individual tumors, including:
- Summary of mutations and copy-number alterations of interest
- Clinical trial information
- TCGA Pathology Reports
- New cancer summary view (Example Endometrial Cancer)
- Updated drug data from KEGG DRUG and NCI Cancer Drugs (aggregated by PiHelper)
#
October 22, 2012
- All data updated to the Broad Firehose run from July 25, 2012.
- COSMIC data added to Mutation Details (via Oncotator).
- All predicted functional impact scores are updated to Mutation Assessor 2.0.
- Users can now base queries on genes in recurrent regions of copy-number alteration (from GISTIC via Firehose).
- The Onco Query Language (OQL) now supports queries for specific mutations or mutation types.
- Data sets added that match the data of all TCGA publications (GBM, ovarian, colorectal, and lung squamous).
#
July 18, 2012
- Mutation data for the TCGA lung squamous cell carcinoma and breast cancer projects (manuscripts in press at Nature).
- All data updated to the latest Broad Firehose run (May 25, 2012).
- Drug information added to the network view (via Drugbank).
- Improved cross-cancer queries: Option to select data types, export of summary graphs.
- Users can now base queries on frequently mutated genes (from MutSig via Firehose).
#
May 16, 2012
- All data updated to the latest Broad Firehose run (March 21, 2012).
- Extended cross-cancer functionality, enabling users to query across all cancer studies in our database.
- New "build a case" functionality, enabling users to generate custom case sets, based on one or more clinical attributes.
- New OncoPrint features, including more compact OncoPrints, and support for RPPA visualization.
#
February 27, 2012
- All data updated to the latest Broad Firehose run (January 24, 2012).
- Validated mutation data for colorectal cancer.
- New feature: Mutation Diagrams that show mutations in the context of protein domains.

#
January 30, 2012
- Updated data for several TCGA cancer studies.
- Some small bug-fixes.
#
December 22, 2011
Fourteen new TCGA cancer studies: This includes complete data for TCGA Colorectal Carcinoma and provisional data for thirteen other cancer types in the TCGA production pipeline. Please note that data from these thirteen new cancer types are provisional, not final and do not yet include mutation data. As per NCI guidelines, preliminary mutation data cannot be redistributed until they have been validated.

Four new data types:
- Reverse-phase protein array (RPPA) data.
- microRNA expression and copy-number (including support for multiple loci)
- RNA-Seq based expression data.
- log2 copy-number data.
Updated TCGA GBM copy-number, expression, and methylation data.
New gene symbol validation service. You can now use gene aliases and/or Entrez Gene IDs within your gene sets.
Links to IGV for visualization of DNA copy-number changes.
Background information from the Sanger Cancer Gene Census.
Two new Tutorials to get you quickly started in using the portal.
#
November 14, 2011
- New and improved mutation details, with sorting and filtering capabilities.
- In collaboration with Bilkent University, we have added a new Network tab to our results pages. The network tab enables users to visualize, analyze and filter cancer genomic data in the context of pathways and interaction networks derived from Pathway Commons.

#
September 3, 2011
- You can now query across different cancer studies (feature available directly from the home page).
- Our MATLAB CGDS Cancer Genomics Toolbox is now available. The toolbox enables you to download data from the cBio Portal, and import it directly into MATLAB.
- The code for the cBio Portal has now been fully open sourced, and made available at Google Code. If you would like to join our open source efforts and make the portal even better, drop us an email.
#
March 2, 2011
New plotting features and other improvements:
- Correlation plots that show the relationship between different data types for individual genes.
- Survival analysis - assess survival differences between altered and non-altered patient sets.
- Updated R Package with support for correlation plots and general improvements for retrieving and accessing data in R data frames.
- The Web Interface now supports basic clinical data, e.g. survival data.
- Networks for pathway analysis are now available for download.

#
December 15, 2010
Several new features, including:
- Redesigned and streamlined user interface, based on user feedback and usability testing.
- Advanced support for gene-specific alterations. For example, users can now view mutations within TP53, and ignore copy number alterations, or only view amplifications of EGFR, and ignore deletions.
- Improved performance.
- Frequently Asked Questions document released.
- Updated
Video Tutorial(update: old link no longer functional. Now see: YouTube
#
November 4, 2010
- Enhanced Oncoprints, enabling users to quickly visualize genomic alterations across many cases. Oncoprints now also work in all major browsers, including Firefox, Chrome, Safari, and Internet Explorer.
- Official release of our Web Interface, enabling programmatic access to all data.
- Official release of our R Package, enabling programmatic access to all data from the R platform for statistical computing.






