#
Using the dataset validator
To facilitate the loading of new studies into its database, cBioPortal provides a set of staging files formats for the various data types. To validate your files you can use the dataset validator script.
#
Running the validator
To run the validator first go to the importer folder
<cbioportal_source_folder>/core/src/main/scripts/importer
and then run the following command:
./validateData.py --helpThis will tell you the parameters you can use:
usage: validateData.py [-h] -s STUDY_DIRECTORY
[-u URL_SERVER | -p PORTAL_INFO_DIR | -n]
[-P PORTAL_PROPERTIES] [-html HTML_TABLE]
[-e ERROR_FILE] [-v] [-r] [-m]
cBioPortal study validator
optional arguments:
-h, --help show this help message and exit
-s STUDY_DIRECTORY, --study_directory STUDY_DIRECTORY
path to directory.
-u URL_SERVER, --url_server URL_SERVER
URL to cBioPortal server. You can set this if your URL
is not http://localhost:8080
-p PORTAL_INFO_DIR, --portal_info_dir PORTAL_INFO_DIR
Path to a directory of cBioPortal info files to be
used instead of contacting a server
-n, --no_portal_checks
Skip tests requiring information from the cBioPortal
installation
-html HTML_TABLE, --html_table HTML_TABLE
path to html report output file
-e ERROR_FILE, --error_file ERROR_FILE
File to which to write line numbers on which errors
were found, for scripts
-v, --verbose report status info messages in addition to errors and
warnings
-r, --relaxed-clinical_definitions
Option to enable relaxed mode for validator when validating
clinical data without header definitions
-m, --strict_maf_checks
Option to enable strict mode for validator when validating
mutation dataFor more information on the --portal_info_dir option, see hg19,
you have to specify the reference_genome field in your meta_study.txt.
For more information, see
When running the validator with parameter -r the validator will run the validation of the clinical data it will ignore all failing checks
about values in the headers of the clinical data file.
When running the validator with parameter -m the validator will run the validation of the specific MAF file checks for the mutation file in strict maf check mode. This means that
when the validator encounters these validation checks it will report them as an error instead of a warning.
#
Example 1: test study_es_0
As an example, you can try the validator with one of the test studies found in <cbioportal_source_folder>/core/src/test/scripts/test_data. Example, assuming port 8080 and using -v option to also see the progress:
./validateData.py -s ../../../test/scripts/test_data/study_es_0/ -u http://localhost:8080 -vResults in:
DEBUG: -: Requesting info from portal at 'http://localhost:8080'
DEBUG: -: Requesting cancer-types from portal at 'http://localhost:8080'
DEBUG: -: Requesting genes from portal at 'http://localhost:8080'
DEBUG: -: Requesting genesets from portal at 'http://localhost:8080'
DEBUG: -: Requesting genesets_version from portal at 'http://localhost:8080'
DEBUG: -: Requesting gene-panels from portal at 'http://localhost:8080'
DEBUG: meta_cancer_type.txt: Starting validation of meta file
INFO: meta_cancer_type.txt: Validation of meta file complete
DEBUG: meta_clinical_patients.txt: Starting validation of meta file
INFO: meta_clinical_patients.txt: Validation of meta file complete
DEBUG: meta_clinical_samples.txt: Starting validation of meta file
INFO: meta_clinical_samples.txt: Validation of meta file complete
DEBUG: meta_cna_discrete.txt: Starting validation of meta file
INFO: meta_cna_discrete.txt: Validation of meta file complete
DEBUG: meta_cna_hg19_seg.txt: Starting validation of meta file
INFO: meta_cna_hg19_seg.txt: Validation of meta file complete
DEBUG: -: Retrieving chromosome lengths from '/home/sander/git/cbioportal/core/src/main/scripts/importer/chromosome_sizes.json'
DEBUG: meta_cna_log2.txt: Starting validation of meta file
INFO: meta_cna_log2.txt: Validation of meta file complete
DEBUG: meta_expression_median.txt: Starting validation of meta file
INFO: meta_expression_median.txt: Validation of meta file complete
DEBUG: meta_expression_median_Zscores.txt: Starting validation of meta file
INFO: meta_expression_median_Zscores.txt: Validation of meta file complete
DEBUG: meta_fusions.txt: Starting validation of meta file
INFO: meta_fusions.txt: Validation of meta file complete
DEBUG: meta_gene_panel_matrix.txt: Starting validation of meta file
INFO: meta_gene_panel_matrix.txt: Validation of meta file complete
DEBUG: meta_gistic_genes_amp.txt: Starting validation of meta file
INFO: meta_gistic_genes_amp.txt: Validation of meta file complete
DEBUG: -: Retrieving chromosome lengths from '/home/sander/git/cbioportal/core/src/main/scripts/importer/chromosome_sizes.json'
DEBUG: meta_gsva_pvalues.txt: Starting validation of meta file
INFO: meta_gsva_pvalues.txt: Validation of meta file complete
DEBUG: meta_gsva_scores.txt: Starting validation of meta file
INFO: meta_gsva_scores.txt: Validation of meta file complete
DEBUG: meta_methylation_hm27.txt: Starting validation of meta file
INFO: meta_methylation_hm27.txt: Validation of meta file complete
DEBUG: meta_mutational_signature.txt: Starting validation of meta file
INFO: meta_mutational_signature.txt: Validation of meta file complete
DEBUG: meta_mutations_extended.txt: Starting validation of meta file
INFO: meta_mutations_extended.txt: Validation of meta file complete
DEBUG: meta_resource_definition.txt: Starting validation of meta file
INFO: meta_resource_definition.txt: Validation of meta file complete
DEBUG: meta_resource_patient.txt: Starting validation of meta file
INFO: meta_resource_patient.txt: Validation of meta file complete
DEBUG: meta_resource_sample.txt: Starting validation of meta file
INFO: meta_resource_sample.txt: Validation of meta file complete
DEBUG: meta_resource_study.txt: Starting validation of meta file
INFO: meta_resource_study.txt: Validation of meta file complete
DEBUG: meta_structural_variants.txt: Starting validation of meta file
INFO: meta_structural_variants.txt: Validation of meta file complete
DEBUG: meta_study.txt: Starting validation of meta file
INFO: meta_study.txt: Validation of meta file complete
DEBUG: -: Study Tag file found. It will be validated.
DEBUG: meta_treatment_ec50.txt: Starting validation of meta file
INFO: meta_treatment_ec50.txt: Validation of meta file complete
DEBUG: meta_treatment_ic50.txt: Starting validation of meta file
INFO: meta_treatment_ic50.txt: Validation of meta file complete
DEBUG: data_cancer_type.txt: Starting validation of file
INFO: data_cancer_type.txt: line 1: New disease type will be added to the portal; value encountered: 'brca-es0'
INFO: data_cancer_type.txt: Validation of file complete
INFO: data_cancer_type.txt: Read 1 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_clinical_samples.txt: Starting validation of file
INFO: data_clinical_samples.txt: Validation of file complete
INFO: data_clinical_samples.txt: Read 847 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_definition.txt: Starting validation of file
INFO: data_resource_definition.txt: Validation of file complete
INFO: data_resource_definition.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_sample.txt: Starting validation of file
INFO: data_resource_sample.txt: Validation of file complete
INFO: data_resource_sample.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: study_tags.yml: Starting validation of study tags file
INFO: study_tags.yml: Validation of study tags file complete.
DEBUG: -: Validating case lists
DEBUG: case_lists/cases_cnaseq.txt: Starting validation of meta file
INFO: case_lists/cases_cnaseq.txt: Validation of meta file complete
DEBUG: case_lists/cases_test.txt: Starting validation of meta file
INFO: case_lists/cases_test.txt: Validation of meta file complete
DEBUG: case_lists/cases_sequenced.txt: Starting validation of meta file
INFO: case_lists/cases_sequenced.txt: Validation of meta file complete
DEBUG: case_lists/cases_custom.txt: Starting validation of meta file
INFO: case_lists/cases_custom.txt: Validation of meta file complete
DEBUG: case_lists/cases_cna.txt: Starting validation of meta file
INFO: case_lists/cases_cna.txt: Validation of meta file complete
INFO: -: Validation of case list folder complete
DEBUG: data_gene_panel_matrix.txt: Starting validation of file
INFO: data_gene_panel_matrix.txt: line 1: This column can be replaced by a 'gene_panel' property in the respective meta file; value encountered: 'gistic'
INFO: data_gene_panel_matrix.txt: Validation of file complete
INFO: data_gene_panel_matrix.txt: Read 21 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_discrete.txt: Starting validation of file
INFO: data_cna_discrete.txt: Validation of file complete
INFO: data_cna_discrete.txt: Read 9 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_clinical_patients.txt: Starting validation of file
INFO: data_clinical_patients.txt: Validation of file complete
INFO: data_clinical_patients.txt: Read 845 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_expression_median.txt: Starting validation of file
INFO: data_expression_median.txt: Validation of file complete
INFO: data_expression_median.txt: Read 7 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_expression_median_Zscores.txt: Starting validation of file
INFO: data_expression_median_Zscores.txt: Validation of file complete
INFO: data_expression_median_Zscores.txt: Read 6 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_fusions.txt: Starting validation of file
INFO: data_fusions.txt: Validation of file complete
INFO: data_fusions.txt: Read 6 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_mutational_signature.txt: Starting validation of file
INFO: data_mutational_signature.txt: Validation of file complete
INFO: data_mutational_signature.txt: Read 62 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_treatment_ec50.txt: Starting validation of file
INFO: data_treatment_ec50.txt: Validation of file complete
INFO: data_treatment_ec50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_treatment_ic50.txt: Starting validation of file
INFO: data_treatment_ic50.txt: Validation of file complete
INFO: data_treatment_ic50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gistic_genes_amp.txt: Starting validation of file
INFO: data_gistic_genes_amp.txt: Validation of file complete
INFO: data_gistic_genes_amp.txt: Read 13 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gsva_pvalues.txt: Starting validation of file
INFO: data_gsva_pvalues.txt: Validation of file complete
INFO: data_gsva_pvalues.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gsva_scores.txt: Starting validation of file
INFO: data_gsva_scores.txt: Validation of file complete
INFO: data_gsva_scores.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_log2.txt: Starting validation of file
INFO: data_cna_log2.txt: Validation of file complete
INFO: data_cna_log2.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_methylation_hm27.txt: Starting validation of file
INFO: data_methylation_hm27.txt: Validation of file complete
INFO: data_methylation_hm27.txt: Read 9 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_mutations_extended.maf: Starting validation of file
INFO: data_mutations_extended.maf: lines [4, 5, 6, (3 more)]: column 164: Values contained in the column cbp_driver_tiers that will appear in the "Mutation Color" menu of the Oncoprint; values encountered: ['Class 2', 'Class 1', 'Class 4', '(1 more)']
INFO: data_mutations_extended.maf: lines [7, 9]: Line will not be loaded due to the variant classification filter. Filtered types: [Silent, Intron, 3'UTR, 3'Flank, 5'UTR, 5'Flank, IGR, RNA]; value encountered: 'Silent'
INFO: data_mutations_extended.maf: Validation of file complete
INFO: data_mutations_extended.maf: Read 35 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_patient.txt: Starting validation of file
INFO: data_resource_patient.txt: Validation of file complete
INFO: data_resource_patient.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_study.txt: Starting validation of file
INFO: data_resource_study.txt: Validation of file complete
INFO: data_resource_study.txt: Read 2 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_hg19.seg: Starting validation of file
INFO: data_cna_hg19.seg: Validation of file complete
INFO: data_cna_hg19.seg: Read 10 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_structural_variants.txt: Starting validation of file
INFO: data_structural_variants.txt: Validation of file complete
INFO: data_structural_variants.txt: Read 46 lines. Lines with warning: 0. Lines with error: 0
INFO: -: Validation complete
Validation of study succeeded.When using the -html option, a report will be generated, which looks like this for the previous example:
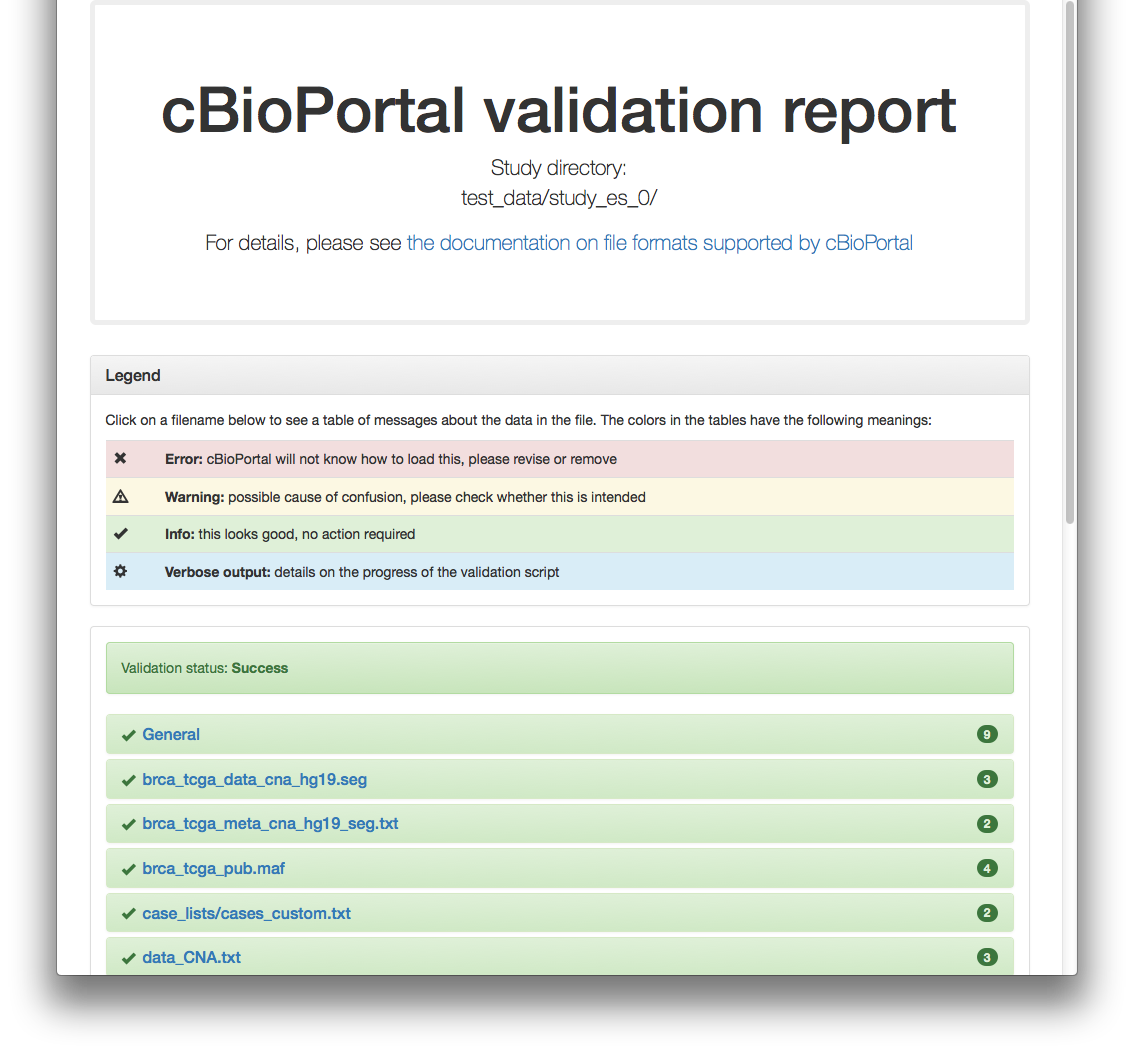
#
Example 2: test study_es_1
More test studies for trying the validator (study_es_1 and study_es_3) are available in <cbioportal_source_folder>/core/src/test/scripts/test_data. Example, assuming port 8080 and using -v option:
./validateData.py -s ../../../test/scripts/test_data/study_es_1/ -u http://localhost:8080 -vResults in:
DEBUG: -: Requesting info from portal at 'http://localhost:8081'
DEBUG: -: Requesting cancer-types from portal at 'http://localhost:8081'
DEBUG: -: Requesting genes from portal at 'http://localhost:8081'
DEBUG: -: Requesting genesets from portal at 'http://localhost:8081'
DEBUG: -: Requesting genesets_version from portal at 'http://localhost:8081'
DEBUG: -: Requesting gene-panels from portal at 'http://localhost:8081'
DEBUG: meta_expression_median.txt: Starting validation of meta file
ERROR: meta_expression_median.txt: Invalid stable id for genetic_alteration_type 'MRNA_EXPRESSION', data_type 'Z-SCORE'; expected one of [mrna_U133_Zscores, rna_seq_mrna_median_Zscores, mrna_median_Zscores, rna_seq_v2_mrna_median_Zscores, rna_seq_v2_mrna_median_normals_Zscores, mirna_median_Zscores, mrna_merged_median_Zscores, mrna_zbynorm, mrna_seq_tpm_Zscores, mrna_seq_cpm_Zscores, rna_seq_mrna_capture_Zscores, mrna_seq_fpkm_capture_Zscores, mrna_seq_fpkm_polya_Zscores, mrna_U133_all_sample_Zscores, mrna_all_sample_Zscores, rna_seq_mrna_median_all_sample_Zscores, mrna_median_all_sample_Zscores, rna_seq_v2_mrna_median_all_sample_Zscores, rna_seq_v2_mrna_median_all_sample_ref_normal_Zscores, mrna_seq_cpm_all_sample_Zscores, mrna_seq_tpm_all_sample_Zscores, rna_seq_mrna_capture_all_sample_Zscores, mrna_seq_fpkm_capture_all_sample_Zscores, mrna_seq_fpkm_polya_all_sample_Zscores]; value encountered: 'mrna'
DEBUG: meta_samples.txt: Starting validation of meta file
WARNING: meta_samples.txt: Unrecognized field in meta file; values encountered: ['show_profile_in_analysis_tab', 'profile_description', 'profile_name']
INFO: meta_samples.txt: Validation of meta file complete
DEBUG: meta_study.txt: Starting validation of meta file
INFO: meta_study.txt: Validation of meta file complete
INFO: meta_study.txt: No reference genome specified -- using default (hg19)
DEBUG: meta_treatment_ec50.txt: Starting validation of meta file
INFO: meta_treatment_ec50.txt: Validation of meta file complete
DEBUG: meta_treatment_ic50.txt: Starting validation of meta file
INFO: meta_treatment_ic50.txt: Validation of meta file complete
DEBUG: data_samples.txt: Starting validation of file
INFO: data_samples.txt: Validation of file complete
INFO: data_samples.txt: Read 831 lines. Lines with warning: 0. Lines with error: 0
DEBUG: -: Validating case lists
DEBUG: case_lists/cases_all.txt: Starting validation of meta file
INFO: case_lists/cases_all.txt: Validation of meta file complete
ERROR: case_lists/cases_all.txt: Sample ID not defined in clinical file; value encountered: 'INVALID-A2-A0T2-01'
INFO: -: Validation of case list folder complete
DEBUG: data_treatment_ec50.txt: Starting validation of file
ERROR: data_treatment_ec50.txt: line 2: column 1: Do not use space in the stable id; value encountered: '17 AAG'
ERROR: data_treatment_ec50.txt: line 7: column 5: Blank cell found in column; value encountered: ''' (in column 'TCGA-A1-A0SB-01')'
INFO: data_treatment_ec50.txt: Validation of file complete
INFO: data_treatment_ec50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 2
DEBUG: data_treatment_ic50.txt: Starting validation of file
ERROR: data_treatment_ic50.txt: line 7: column 5: Blank cell found in column; value encountered: ''' (in column 'TCGA-A1-A0SB-01')'
INFO: data_treatment_ic50.txt: Validation of file complete
INFO: data_treatment_ic50.txt: Read 10 lines. Lines with warning: 0. Lines with error: 1
INFO: -: Validation complete
Validation of study failed.And respective HTML report:
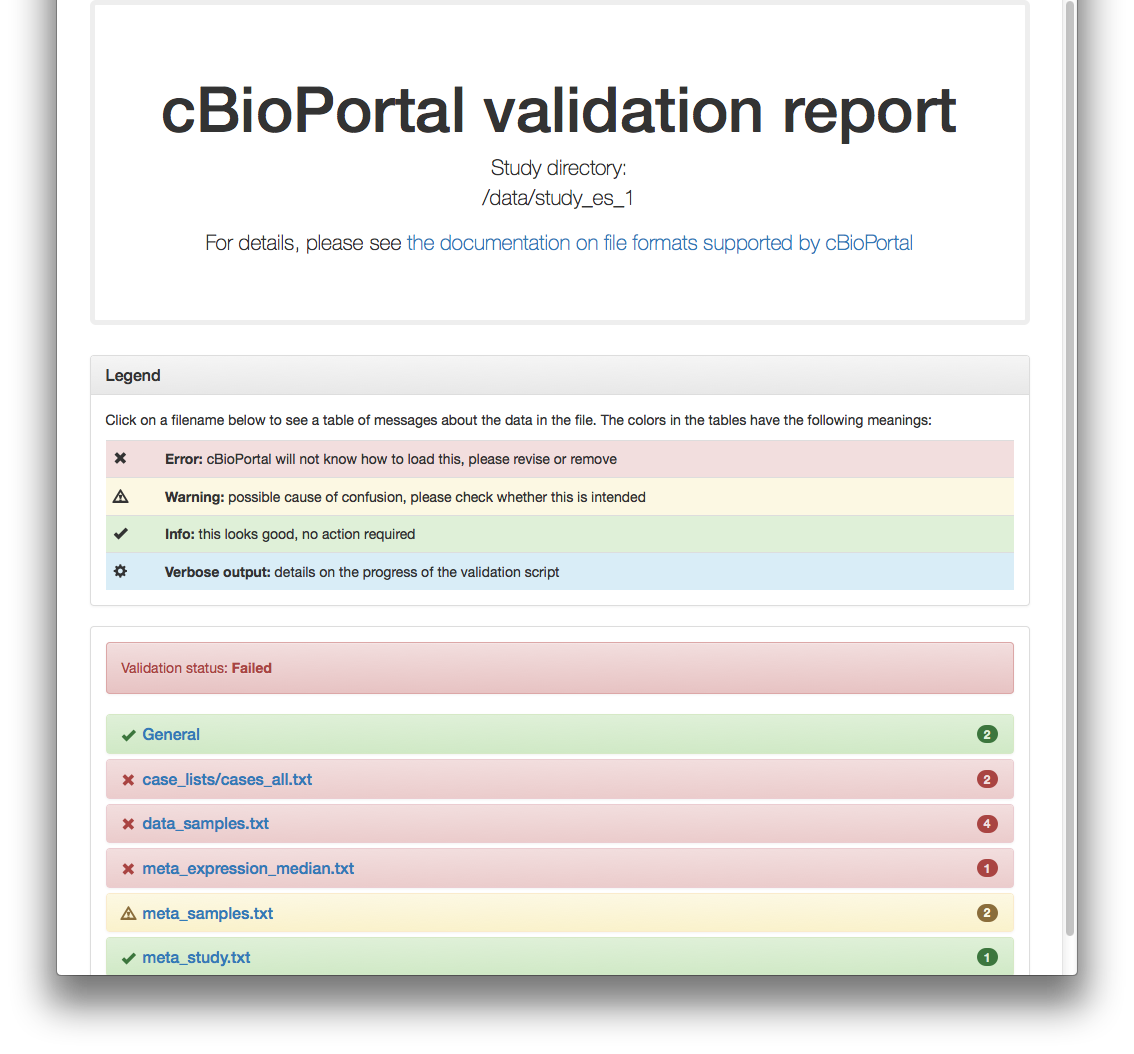
#
Offline validation
The validation script can be used offline, without connecting to a cBioPortal server. The tests that depend on information specific to the portal (which clinical attributes and cancer types have been previously defined, and which Entrez gene identifiers and corresponding symbols are supported), will instead be read from a folder with .json files generated from the portal.
#
Example 3: validation with a portal info folder
To run the validator with a folder of portal information files, add the -p/--portal_info_dir option to the command line, followed by the path to the folder:
./validateData.py -s ../../../test/scripts/test_data/study_es_0/ -p ../../../test/scripts/test_data/api_json_system_tests/ -vDEBUG: -: Reading portal information from ../../../test/scripts/test_data/api_json_system_tests/cancer-types.json
DEBUG: -: Reading portal information from ../../../test/scripts/test_data/api_json_system_tests/genes.json
DEBUG: -: Reading portal information from ../../../test/scripts/test_data/api_json_system_tests/genesets.json
DEBUG: -: Reading portal information from ../../../test/scripts/test_data/api_json_system_tests/genesets_version.json
DEBUG: -: Reading portal information from ../../../test/scripts/test_data/api_json_system_tests/gene-panels.json
DEBUG: meta_cancer_type.txt: Starting validation of meta file
INFO: meta_cancer_type.txt: Validation of meta file complete
DEBUG: meta_clinical_patients.txt: Starting validation of meta file
INFO: meta_clinical_patients.txt: Validation of meta file complete
DEBUG: meta_clinical_samples.txt: Starting validation of meta file
INFO: meta_clinical_samples.txt: Validation of meta file complete
DEBUG: meta_cna_discrete.txt: Starting validation of meta file
INFO: meta_cna_discrete.txt: Validation of meta file complete
DEBUG: meta_cna_hg19_seg.txt: Starting validation of meta file
INFO: meta_cna_hg19_seg.txt: Validation of meta file complete
DEBUG: -: Retrieving chromosome lengths from '/home/sander/git/cbioportal/core/src/main/scripts/importer/chromosome_sizes.json'
DEBUG: meta_cna_log2.txt: Starting validation of meta file
INFO: meta_cna_log2.txt: Validation of meta file complete
DEBUG: meta_expression_median.txt: Starting validation of meta file
INFO: meta_expression_median.txt: Validation of meta file complete
DEBUG: meta_expression_median_Zscores.txt: Starting validation of meta file
INFO: meta_expression_median_Zscores.txt: Validation of meta file complete
DEBUG: meta_fusions.txt: Starting validation of meta file
INFO: meta_fusions.txt: Validation of meta file complete
DEBUG: meta_gene_panel_matrix.txt: Starting validation of meta file
INFO: meta_gene_panel_matrix.txt: Validation of meta file complete
DEBUG: meta_gistic_genes_amp.txt: Starting validation of meta file
INFO: meta_gistic_genes_amp.txt: Validation of meta file complete
DEBUG: -: Retrieving chromosome lengths from '/home/sander/git/cbioportal/core/src/main/scripts/importer/chromosome_sizes.json'
DEBUG: meta_gsva_pvalues.txt: Starting validation of meta file
INFO: meta_gsva_pvalues.txt: Validation of meta file complete
DEBUG: meta_gsva_scores.txt: Starting validation of meta file
INFO: meta_gsva_scores.txt: Validation of meta file complete
DEBUG: meta_methylation_hm27.txt: Starting validation of meta file
INFO: meta_methylation_hm27.txt: Validation of meta file complete
DEBUG: meta_mutational_signature.txt: Starting validation of meta file
INFO: meta_mutational_signature.txt: Validation of meta file complete
DEBUG: meta_mutations_extended.txt: Starting validation of meta file
INFO: meta_mutations_extended.txt: Validation of meta file complete
DEBUG: meta_resource_definition.txt: Starting validation of meta file
INFO: meta_resource_definition.txt: Validation of meta file complete
DEBUG: meta_resource_patient.txt: Starting validation of meta file
INFO: meta_resource_patient.txt: Validation of meta file complete
DEBUG: meta_resource_sample.txt: Starting validation of meta file
INFO: meta_resource_sample.txt: Validation of meta file complete
DEBUG: meta_resource_study.txt: Starting validation of meta file
INFO: meta_resource_study.txt: Validation of meta file complete
DEBUG: meta_structural_variants.txt: Starting validation of meta file
INFO: meta_structural_variants.txt: Validation of meta file complete
DEBUG: meta_study.txt: Starting validation of meta file
INFO: meta_study.txt: Validation of meta file complete
DEBUG: -: Study Tag file found. It will be validated.
DEBUG: meta_treatment_ec50.txt: Starting validation of meta file
INFO: meta_treatment_ec50.txt: Validation of meta file complete
DEBUG: meta_treatment_ic50.txt: Starting validation of meta file
INFO: meta_treatment_ic50.txt: Validation of meta file complete
DEBUG: data_cancer_type.txt: Starting validation of file
INFO: data_cancer_type.txt: Validation of file complete
INFO: data_cancer_type.txt: Read 1 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_clinical_samples.txt: Starting validation of file
INFO: data_clinical_samples.txt: Validation of file complete
INFO: data_clinical_samples.txt: Read 847 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_definition.txt: Starting validation of file
INFO: data_resource_definition.txt: Validation of file complete
INFO: data_resource_definition.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_sample.txt: Starting validation of file
INFO: data_resource_sample.txt: Validation of file complete
INFO: data_resource_sample.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: study_tags.yml: Starting validation of study tags file
INFO: study_tags.yml: Validation of study tags file complete.
DEBUG: -: Validating case lists
DEBUG: case_lists/cases_cnaseq.txt: Starting validation of meta file
INFO: case_lists/cases_cnaseq.txt: Validation of meta file complete
DEBUG: case_lists/cases_test.txt: Starting validation of meta file
INFO: case_lists/cases_test.txt: Validation of meta file complete
DEBUG: case_lists/cases_sequenced.txt: Starting validation of meta file
INFO: case_lists/cases_sequenced.txt: Validation of meta file complete
DEBUG: case_lists/cases_custom.txt: Starting validation of meta file
INFO: case_lists/cases_custom.txt: Validation of meta file complete
DEBUG: case_lists/cases_cna.txt: Starting validation of meta file
INFO: case_lists/cases_cna.txt: Validation of meta file complete
INFO: -: Validation of case list folder complete
DEBUG: data_gene_panel_matrix.txt: Starting validation of file
INFO: data_gene_panel_matrix.txt: line 1: This column can be replaced by a 'gene_panel' property in the respective meta file; value encountered: 'gistic'
INFO: data_gene_panel_matrix.txt: Validation of file complete
INFO: data_gene_panel_matrix.txt: Read 21 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_discrete.txt: Starting validation of file
INFO: data_cna_discrete.txt: Validation of file complete
INFO: data_cna_discrete.txt: Read 9 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_clinical_patients.txt: Starting validation of file
INFO: data_clinical_patients.txt: Validation of file complete
INFO: data_clinical_patients.txt: Read 845 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_expression_median.txt: Starting validation of file
INFO: data_expression_median.txt: Validation of file complete
INFO: data_expression_median.txt: Read 7 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_expression_median_Zscores.txt: Starting validation of file
INFO: data_expression_median_Zscores.txt: Validation of file complete
INFO: data_expression_median_Zscores.txt: Read 6 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_fusions.txt: Starting validation of file
INFO: data_fusions.txt: Validation of file complete
INFO: data_fusions.txt: Read 6 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_mutational_signature.txt: Starting validation of file
INFO: data_mutational_signature.txt: Validation of file complete
INFO: data_mutational_signature.txt: Read 62 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_treatment_ec50.txt: Starting validation of file
INFO: data_treatment_ec50.txt: Validation of file complete
INFO: data_treatment_ec50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_treatment_ic50.txt: Starting validation of file
INFO: data_treatment_ic50.txt: Validation of file complete
INFO: data_treatment_ic50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gistic_genes_amp.txt: Starting validation of file
INFO: data_gistic_genes_amp.txt: Validation of file complete
INFO: data_gistic_genes_amp.txt: Read 13 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gsva_pvalues.txt: Starting validation of file
INFO: data_gsva_pvalues.txt: Validation of file complete
INFO: data_gsva_pvalues.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gsva_scores.txt: Starting validation of file
INFO: data_gsva_scores.txt: Validation of file complete
INFO: data_gsva_scores.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_log2.txt: Starting validation of file
INFO: data_cna_log2.txt: Validation of file complete
INFO: data_cna_log2.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_methylation_hm27.txt: Starting validation of file
INFO: data_methylation_hm27.txt: Validation of file complete
INFO: data_methylation_hm27.txt: Read 9 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_mutations_extended.maf: Starting validation of file
INFO: data_mutations_extended.maf: lines [4, 5, 6, (3 more)]: column 164: Values contained in the column cbp_driver_tiers that will appear in the "Mutation Color" menu of the Oncoprint; values encountered: ['Class 2', 'Class 1', 'Class 4', '(1 more)']
INFO: data_mutations_extended.maf: lines [7, 9]: Line will not be loaded due to the variant classification filter. Filtered types: [Silent, Intron, 3'UTR, 3'Flank, 5'UTR, 5'Flank, IGR, RNA]; value encountered: 'Silent'
INFO: data_mutations_extended.maf: Validation of file complete
INFO: data_mutations_extended.maf: Read 35 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_patient.txt: Starting validation of file
INFO: data_resource_patient.txt: Validation of file complete
INFO: data_resource_patient.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_study.txt: Starting validation of file
INFO: data_resource_study.txt: Validation of file complete
INFO: data_resource_study.txt: Read 2 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_hg19.seg: Starting validation of file
INFO: data_cna_hg19.seg: Validation of file complete
INFO: data_cna_hg19.seg: Read 10 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_structural_variants.txt: Starting validation of file
INFO: data_structural_variants.txt: Validation of file complete
INFO: data_structural_variants.txt: Read 46 lines. Lines with warning: 0. Lines with error: 0
INFO: -: Validation complete
Validation of study succeeded.
#
Example 4: generating the portal info folder
The portal information files can be generated on the server, using the dumpPortalInfo script. Go to <cbioportal_source_folder>/core/src/main/scripts, make sure the environment variables $JAVA_HOME and $PORTAL_HOME are set, and run dumpPortalInfo.pl with the name of the directory you want to create:
export JAVA_HOME='/usr/lib/jvm/default-java'
export PORTAL_HOME=<cbioportal_configuration_folder>
./dumpPortalInfo.pl /home/johndoe/my_portal_info_folder/
#
Example 5: validating without portal-specific information
Alternatively, you can run the validation script with the -n/--no_portal_checks flag to entirely skip checks relating to installation-specific metadata. Be warned that files succeeding this validation may still fail to load (correctly).
./validateData.py -s ../../../test/scripts/test_data/study_es_0/ -n -vWARNING: -: Skipping validations relating to cancer types defined in the portal
WARNING: -: Skipping validations relating to gene identifiers and aliases defined in the portal
WARNING: -: Skipping validations relating to gene set identifiers
WARNING: -: Skipping validations relating to gene panel identifiers
DEBUG: meta_cancer_type.txt: Starting validation of meta file
INFO: meta_cancer_type.txt: Validation of meta file complete
DEBUG: meta_clinical_patients.txt: Starting validation of meta file
INFO: meta_clinical_patients.txt: Validation of meta file complete
DEBUG: meta_clinical_samples.txt: Starting validation of meta file
INFO: meta_clinical_samples.txt: Validation of meta file complete
DEBUG: meta_cna_discrete.txt: Starting validation of meta file
INFO: meta_cna_discrete.txt: Validation of meta file complete
DEBUG: meta_cna_hg19_seg.txt: Starting validation of meta file
INFO: meta_cna_hg19_seg.txt: Validation of meta file complete
DEBUG: -: Retrieving chromosome lengths from '/home/sander/git/cbioportal/core/src/main/scripts/importer/chromosome_sizes.json'
DEBUG: meta_cna_log2.txt: Starting validation of meta file
INFO: meta_cna_log2.txt: Validation of meta file complete
DEBUG: meta_expression_median.txt: Starting validation of meta file
INFO: meta_expression_median.txt: Validation of meta file complete
DEBUG: meta_expression_median_Zscores.txt: Starting validation of meta file
INFO: meta_expression_median_Zscores.txt: Validation of meta file complete
DEBUG: meta_fusions.txt: Starting validation of meta file
INFO: meta_fusions.txt: Validation of meta file complete
DEBUG: meta_gene_panel_matrix.txt: Starting validation of meta file
INFO: meta_gene_panel_matrix.txt: Validation of meta file complete
DEBUG: meta_gistic_genes_amp.txt: Starting validation of meta file
INFO: meta_gistic_genes_amp.txt: Validation of meta file complete
DEBUG: -: Retrieving chromosome lengths from '/home/sander/git/cbioportal/core/src/main/scripts/importer/chromosome_sizes.json'
DEBUG: meta_gsva_pvalues.txt: Starting validation of meta file
INFO: meta_gsva_pvalues.txt: Validation of meta file complete
DEBUG: meta_gsva_scores.txt: Starting validation of meta file
INFO: meta_gsva_scores.txt: Validation of meta file complete
DEBUG: meta_methylation_hm27.txt: Starting validation of meta file
INFO: meta_methylation_hm27.txt: Validation of meta file complete
DEBUG: meta_mutational_signature.txt: Starting validation of meta file
INFO: meta_mutational_signature.txt: Validation of meta file complete
DEBUG: meta_mutations_extended.txt: Starting validation of meta file
INFO: meta_mutations_extended.txt: Validation of meta file complete
DEBUG: meta_resource_definition.txt: Starting validation of meta file
INFO: meta_resource_definition.txt: Validation of meta file complete
DEBUG: meta_resource_patient.txt: Starting validation of meta file
INFO: meta_resource_patient.txt: Validation of meta file complete
DEBUG: meta_resource_sample.txt: Starting validation of meta file
INFO: meta_resource_sample.txt: Validation of meta file complete
DEBUG: meta_resource_study.txt: Starting validation of meta file
INFO: meta_resource_study.txt: Validation of meta file complete
DEBUG: meta_structural_variants.txt: Starting validation of meta file
INFO: meta_structural_variants.txt: Validation of meta file complete
DEBUG: meta_study.txt: Starting validation of meta file
INFO: meta_study.txt: Validation of meta file complete
DEBUG: -: Study Tag file found. It will be validated.
DEBUG: meta_treatment_ec50.txt: Starting validation of meta file
INFO: meta_treatment_ec50.txt: Validation of meta file complete
DEBUG: meta_treatment_ic50.txt: Starting validation of meta file
INFO: meta_treatment_ic50.txt: Validation of meta file complete
DEBUG: data_cancer_type.txt: Starting validation of file
INFO: data_cancer_type.txt: Validation of file complete
INFO: data_cancer_type.txt: Read 1 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_clinical_samples.txt: Starting validation of file
INFO: data_clinical_samples.txt: Validation of file complete
INFO: data_clinical_samples.txt: Read 847 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_definition.txt: Starting validation of file
INFO: data_resource_definition.txt: Validation of file complete
INFO: data_resource_definition.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_sample.txt: Starting validation of file
INFO: data_resource_sample.txt: Validation of file complete
INFO: data_resource_sample.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: study_tags.yml: Starting validation of study tags file
INFO: study_tags.yml: Validation of study tags file complete.
DEBUG: -: Validating case lists
DEBUG: case_lists/cases_cnaseq.txt: Starting validation of meta file
INFO: case_lists/cases_cnaseq.txt: Validation of meta file complete
DEBUG: case_lists/cases_test.txt: Starting validation of meta file
INFO: case_lists/cases_test.txt: Validation of meta file complete
DEBUG: case_lists/cases_sequenced.txt: Starting validation of meta file
INFO: case_lists/cases_sequenced.txt: Validation of meta file complete
DEBUG: case_lists/cases_custom.txt: Starting validation of meta file
INFO: case_lists/cases_custom.txt: Validation of meta file complete
DEBUG: case_lists/cases_cna.txt: Starting validation of meta file
INFO: case_lists/cases_cna.txt: Validation of meta file complete
INFO: -: Validation of case list folder complete
DEBUG: data_gene_panel_matrix.txt: Starting validation of file
INFO: data_gene_panel_matrix.txt: line 1: This column can be replaced by a 'gene_panel' property in the respective meta file; value encountered: 'gistic'
INFO: data_gene_panel_matrix.txt: Validation of file complete
INFO: data_gene_panel_matrix.txt: Read 21 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_discrete.txt: Starting validation of file
INFO: data_cna_discrete.txt: Validation of file complete
INFO: data_cna_discrete.txt: Read 9 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_clinical_patients.txt: Starting validation of file
INFO: data_clinical_patients.txt: Validation of file complete
INFO: data_clinical_patients.txt: Read 845 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_expression_median.txt: Starting validation of file
INFO: data_expression_median.txt: Validation of file complete
INFO: data_expression_median.txt: Read 7 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_expression_median_Zscores.txt: Starting validation of file
INFO: data_expression_median_Zscores.txt: Validation of file complete
INFO: data_expression_median_Zscores.txt: Read 6 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_fusions.txt: Starting validation of file
INFO: data_fusions.txt: Validation of file complete
INFO: data_fusions.txt: Read 6 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_mutational_signature.txt: Starting validation of file
INFO: data_mutational_signature.txt: Validation of file complete
INFO: data_mutational_signature.txt: Read 62 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_treatment_ec50.txt: Starting validation of file
INFO: data_treatment_ec50.txt: Validation of file complete
INFO: data_treatment_ec50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_treatment_ic50.txt: Starting validation of file
INFO: data_treatment_ic50.txt: Validation of file complete
INFO: data_treatment_ic50.txt: Read 11 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gistic_genes_amp.txt: Starting validation of file
INFO: data_gistic_genes_amp.txt: Validation of file complete
INFO: data_gistic_genes_amp.txt: Read 13 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gsva_pvalues.txt: Starting validation of file
INFO: data_gsva_pvalues.txt: Validation of file complete
INFO: data_gsva_pvalues.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_gsva_scores.txt: Starting validation of file
INFO: data_gsva_scores.txt: Validation of file complete
INFO: data_gsva_scores.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_log2.txt: Starting validation of file
INFO: data_cna_log2.txt: Validation of file complete
INFO: data_cna_log2.txt: Read 8 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_methylation_hm27.txt: Starting validation of file
INFO: data_methylation_hm27.txt: Validation of file complete
INFO: data_methylation_hm27.txt: Read 9 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_mutations_extended.maf: Starting validation of file
INFO: data_mutations_extended.maf: lines [4, 5, 6, (3 more)]: column 164: Values contained in the column cbp_driver_tiers that will appear in the "Mutation Color" menu of the Oncoprint; values encountered: ['Class 2', 'Class 1', 'Class 4', '(1 more)']
INFO: data_mutations_extended.maf: lines [7, 9]: Line will not be loaded due to the variant classification filter. Filtered types: [Silent, Intron, 3'UTR, 3'Flank, 5'UTR, 5'Flank, IGR, RNA]; value encountered: 'Silent'
INFO: data_mutations_extended.maf: Validation of file complete
INFO: data_mutations_extended.maf: Read 35 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_patient.txt: Starting validation of file
INFO: data_resource_patient.txt: Validation of file complete
INFO: data_resource_patient.txt: Read 4 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_resource_study.txt: Starting validation of file
INFO: data_resource_study.txt: Validation of file complete
INFO: data_resource_study.txt: Read 2 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_cna_hg19.seg: Starting validation of file
INFO: data_cna_hg19.seg: Validation of file complete
INFO: data_cna_hg19.seg: Read 10 lines. Lines with warning: 0. Lines with error: 0
DEBUG: data_structural_variants.txt: Starting validation of file
INFO: data_structural_variants.txt: Validation of file complete
INFO: data_structural_variants.txt: Read 46 lines. Lines with warning: 0. Lines with error: 0
INFO: -: Validation complete
Validation of study succeeded with warnings.
#
Validation of non-human data
When importing a study with a reference genome other than hg19/GRCh37, this should be specified in the meta_study.txt file, next to the reference_genome field. Supported values are hg19, hg38 and mm10.
cBioPortal is gradually introducing support for mouse. If you want to load mouse studies and you have to set up your database for mouse.
As an example, the command for the mouse example using the three parameters is given:
./validateData.py -s ../../../test/scripts/test_data/study_es_0/ -P ../../../../../src/main/resources/application.properties -u http://localhost:8080 -v
#
Running the validator for multiple studies
The importer folder <cbioportal_source_folder>/core/src/main/scripts/importer also contains a script for running the validator for multiple studies:
./validateStudies.py --helpThe following parameters can be used:
usage: validateStudies.py [-h] [-d ROOT_DIRECTORY] [-l LIST_OF_STUDIES]
[-html HTML_FOLDER]
[-u URL_SERVER | -p PORTAL_INFO_DIR | -n]
[-P PORTAL_PROPERTIES] [-m]
Wrapper where cBioPortal study validator is run for multiple studies
optional arguments:
-h, --help show this help message and exit
-d ROOT_DIRECTORY, --root-directory ROOT_DIRECTORY
Path to directory with all studies that should be
validated
-l LIST_OF_STUDIES, --list-of-studies LIST_OF_STUDIES
List with paths of studies which should be validated
-html HTML_FOLDER, --html-folder HTML_FOLDER
Path to folder for output HTML reports
-u URL_SERVER, --url_server URL_SERVER
URL to cBioPortal server. You can set this if your URL
is not http://localhost:8080
-p PORTAL_INFO_DIR, --portal_info_dir PORTAL_INFO_DIR
Path to a directory of cBioPortal info files to be
used instead of contacting a server
-n, --no_portal_checks
Skip tests requiring information from the cBioPortal
installation
-m, --strict_maf_checks
Option to enable strict mode for validator when
validating mutation dataParameters --url_server, --portal_info_dir, --no_portal_checks and --portal_properties are equal to the parameters with the same name in validateData.py. The script will save a log file with validation output (log-validate-studies.txt) and output the validation status from the input studies:
=== Validating study ../../../test/scripts/test_data/study_es_0
Result: VALID (WITH WARNINGS)
=== Validating study ../../../test/scripts/test_data/study_es_1
Result: INVALID
=== Validating study ../../../test/scripts/test_data/study_es_invalid
directory cannot be found: ../../../test/scripts/test_data/study_es_invalid
Result: INVALID (PROBLEMS OCCURRED)
#
Example 1: Root directory parameter
Validation can be run for all studies in a certain directory by using the --root-directory parameter. The script will append each folder in the root directory to the study list to validate:
./validateStudies.py -d ../../../test/scripts/test_data/
#
Example 2: List of studies parameter
Validation can also be run for specific studies by using the --list-of-studies parameter. The paths to the different studies can be defined and seperated by a comma:
./validateStudies.py -l ../../../test/scripts/test_data/study_es_0,../../../test/scripts/test_data/study_es_1
#
Example 3: Combination root directory and list of studies parameter
Validation can also be run on specific studies in a certain directory by combining the --root-directory and --list-of-studies parameter:
./validateStudies.py -d ../../../test/scripts/test_data/ -l study_es_0,study_es_1
#
Example 4: HTML folder parameter
When HTML validation reports are desired, an output folder for these HTML files can be specified. This folder does not have to exist, the script can create the folder. The HTML validation reports will get the following name: <study_name>-validation.html. To create HTML validation reports for each study the --html-folder parameter needs to be defined:
./validateStudies.py -d ../../../test/scripts/test_data/ -l study_es_0,study_es_1 -html ../../../test/scripts/test_data/validation-reports